5T5B
 
 | |
3TWC
 
 | |
3TYJ
 
 | | Bacillus collagen-like protein of anthracis P159S mutant | | Descriptor: | BclA protein | | Authors: | Kirchdoerfer, R.N, Herrin, B.R, Han, B.W, Turnbough Jr, C.L, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2011-09-26 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores.
Structure, 20, 2012
|
|
5T29
 
 | | Crystal structure of 10E8 Fab light chain mutant3, against the MPER region of the HIV-1 Env, in complex with the MPER epitope scaffold T117v2 | | Descriptor: | 10E8 EPITOPE SCAFFOLD T117V2, Antibody 10E8 FAB HEAVY CHAIN, Antibody 10E8 FAB LIGHT CHAIN, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2016-08-23 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design.
PLoS Pathog., 13, 2017
|
|
3KU6
 
 | | Crystal structure of a H2N2 influenza virus hemagglutinin, 226L/228G | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-19 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic.
J.Virol., 84, 2010
|
|
3MJ9
 
 | |
3LZG
 
 | | Crystal structure of a 2009 H1N1 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2010-03-01 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus.
Science, 328, 2010
|
|
2GAR
 
 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
5T3S
 
 | | HIV gp140 trimer MD39-10MUTA in complex with Fabs PGT124 and 35022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-08-26 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies.
Immunity, 45, 2016
|
|
3MUG
 
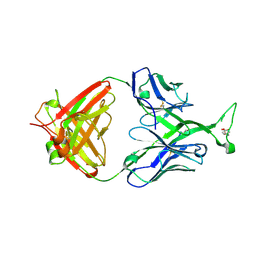 | | Crystal structure of human Fab PG16, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody PG16 Heavy Chain, ... | | Authors: | Pejchal, R, Walker, L.M, Burton, D.R, Wilson, I.A. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MTX
 
 | | Crystal structure of chicken MD-1 | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, GLYCEROL, Protein MD-1, ... | | Authors: | Yoon, S.I, Hong, M, Han, G.W, Wilson, I.A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of soluble MD-1 and its interaction with lipid IVa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MUH
 
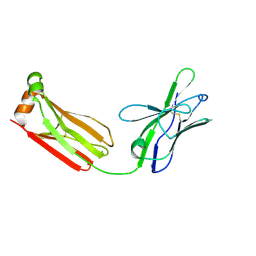 | | Crystal structure of PG9 light chain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody PG9 light chain | | Authors: | Pejchal, R, Walker, L.M, Burton, D.R, Wilson, I.A. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5TH0
 
 | |
5THC
 
 | |
5T6L
 
 | | Crystal structure of 10E8 Fab in complex with the MPER epitope scaffold T117v2 | | Descriptor: | 10E8 EPITOPE SCAFFOLD T117V2, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Antibody 10E8 FAB HEAVY CHAIN, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2016-09-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design.
PLoS Pathog., 13, 2017
|
|
5TGO
 
 | |
2IAD
 
 | |
5T6N
 
 | |
5T80
 
 | |
5TGU
 
 | |
5TFW
 
 | | Crystal structure of 10E8 Fab light chain mutant2 against the MPER region of the HIV-1 Env, in complex with T117v2 epitope scaffold | | Descriptor: | 1,2-ETHANEDIOL, 10E8 EPITOPE SCAFFOLD T117V2, Antibody 10E8 FAB HEAVY CHAIN, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2016-09-26 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design.
PLoS Pathog., 13, 2017
|
|
3TCT
 
 | | Structure of wild-type TTR in complex with tafamidis | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Connelly, S, Kelly, J.W, Wilson, I.A. | | Deposit date: | 2011-08-09 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5T85
 
 | |
5T6S
 
 | |
3MU3
 
 | | Crystal structure of chicken MD-1 complexed with lipid IVa | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-deoxy-3-O-[(3R)-3-hydroxytetradecanoyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-4-O-phosphono-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Yoon, S.I, Hong, M, Han, G.W, Wilson, I.A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of soluble MD-1 and its interaction with lipid IVa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
