4F8R
 
 | |
3PX6
 
 | |
3PV8
 
 | |
3PX0
 
 | |
3PS5
 
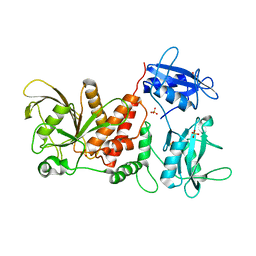 | | Crystal structure of the full-length Human Protein Tyrosine Phosphatase SHP-1 | | 分子名称: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 6 | | 著者 | Wang, W, Liu, L, Song, X, Mo, Y, Komma, C, Bellamy, H.D, Zhao, Z.J, Zhou, G.W. | | 登録日 | 2010-11-30 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of human protein tyrosine phosphatase SHP-1 in the open conformation.
J.Cell.Biochem., 112, 2011
|
|
3PX4
 
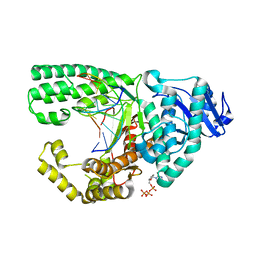 | |
8GY6
 
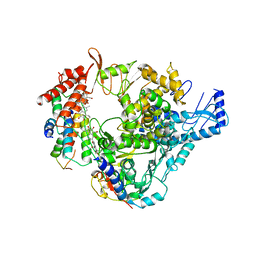 | |
4L67
 
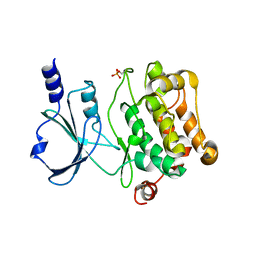 | | Crystal Structure of Catalytic Domain of PAK4 | | 分子名称: | Serine/threonine-protein kinase PAK 4 | | 著者 | Wang, W, Song, J. | | 登録日 | 2013-06-12 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | NMR binding and crystal structure reveal that intrinsically-unstructured regulatory domain auto-inhibits PAK4 by a mechanism different for that of PAK1
Biochem.Biophys.Res.Commun., 438, 2013
|
|
8XZG
 
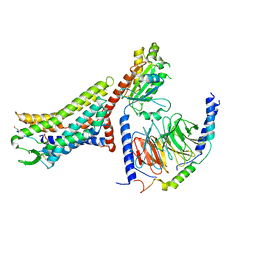 | | Cryo-EM structure of the [Pyr1]-apelin-13-bound human APLNR-Gi complex | | 分子名称: | Apelin receptor, Apelin-13, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Wang, W, Ji, S, Zhang, Y. | | 登録日 | 2024-01-21 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
7XUR
 
 | |
8GYN
 
 | | zebrafish TIPE1 strucutre in complex with PE | | 分子名称: | Tumor necrosis factor alpha-induced protein 8-like protein 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | 著者 | Wang, W, Cao, S.J. | | 登録日 | 2022-09-23 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Structural insight into TIPE1 functioning as a lipid transfer protein.
J.Biomol.Struct.Dyn., 41, 2023
|
|
7TNY
 
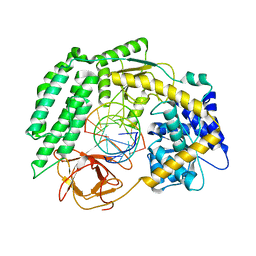 | | Cryo-EM structure of RIG-I in complex with p2dsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, ZINC ION, p2dsRNA | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNX
 
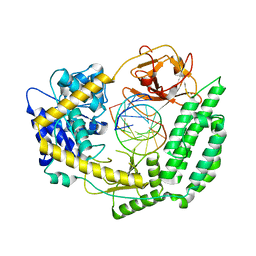 | | Cryo-EM structure of RIG-I in complex with p3dsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, ZINC ION, p3dsRNAa, ... | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TO2
 
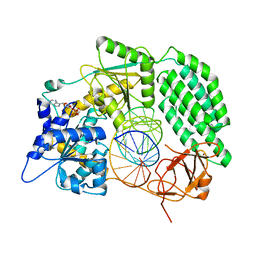 | |
7TO1
 
 | |
7TO0
 
 | | Cryo-EM structure of RIG-I in complex with OHdsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, OHdsRNA, ZINC ION | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNZ
 
 | | Cryo-EM structure of RIG-I in complex with p1dsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, ZINC ION, p1dsRNA | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
8K8T
 
 | | Structure of CUL3-RBX1-KLHL22 complex | | 分子名称: | Cullin-3, Kelch-like protein 22 | | 著者 | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | 登録日 | 2023-07-31 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8K9I
 
 | | Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | 著者 | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | 登録日 | 2023-08-01 | | 公開日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
4FXW
 
 | | Structure of phosphorylated SF1 complex with U2AF65-UHM domain | | 分子名称: | SULFATE ION, Splicing factor 1, Splicing factor U2AF 65 kDa subunit | | 著者 | Wang, W, Bauer, W.J, Wedekind, J.E, Kielkopf, C.L. | | 登録日 | 2012-07-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structure of Phosphorylated SF1 Bound to U2AF(65) in an Essential Splicing Factor Complex.
Structure, 21, 2013
|
|
2MBE
 
 | |
5WVM
 
 | | Crystal structure of baeS cocrystallized with 2 mM indole | | 分子名称: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | 著者 | Wang, W, Zhang, Y, Rang, T, Xu, D. | | 登録日 | 2016-12-26 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
5WVN
 
 | | Crystal structure of MBS-BaeS fusion protein | | 分子名称: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | 著者 | Wang, W, Zhang, Y, Ran, T, Xu, D. | | 登録日 | 2016-12-26 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
3TAR
 
 | |
3SSK
 
 | |
