7XD8
 
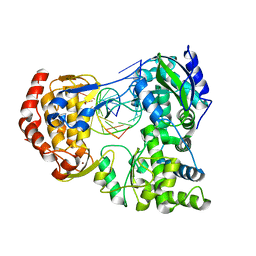 | | Crystal Structure of Dengue Virus Serotype 2 (DENV2) Polymerase Elongation Complex (Native Form) | | 分子名称: | GLYCEROL, NS5, RNA (30-mer), ... | | 著者 | Wu, J, Wang, X, Gong, P. | | 登録日 | 2022-03-26 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural basis of transition from initiation to elongation in de novo viral RNA-dependent RNA polymerases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5ZRV
 
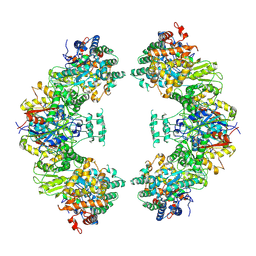 | | Structure of human mitochondrial trifunctional protein, octamer | | 分子名称: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | 著者 | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | 登録日 | 2018-04-25 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (7.7 Å) | | 主引用文献 | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4GHQ
 
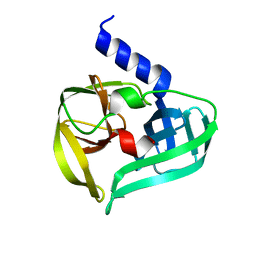 | | Crystal structure of EV71 3C proteinase | | 分子名称: | 3C proteinase | | 著者 | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | 登録日 | 2012-08-08 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | 分子名称: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | 著者 | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | 登録日 | 2012-08-08 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5K0X
 
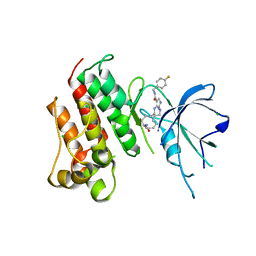 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2541 | | 分子名称: | (7S)-7-amino-N-[(4-fluorophenyl)methyl]-8-oxo-2,9,16,18,21-pentaazabicyclo[15.3.1]henicosa-1(21),17,19-triene-20-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | 著者 | McIver, A.L, Zhang, W, Liu, Q, Jiang, X, Stashko, M.A, Nichols, J, Miley, M.J, Norris-Drouin, J, Machius, M, DeRyckere, D, Wood, E, Graham, D.K, Earp, H.S, Kireev, D, Frye, S.V, Wang, X. | | 登録日 | 2016-05-17 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.231 Å) | | 主引用文献 | Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors.
ChemMedChem, 12, 2017
|
|
7XYO
 
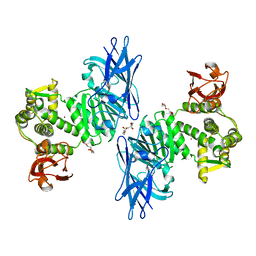 | | Crystal Structure of a M61 aminopeptidase family member from Myxococcus fulvus | | 分子名称: | Aminopeptidase M61, HEXAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | 著者 | Chen, X, Wang, X, Huo, L, Wu, D. | | 登録日 | 2022-06-02 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery and Characterization of a Myxobacterial Lanthipeptide with Unique Biosynthetic Features and Anti-inflammatory Activity.
J.Am.Chem.Soc., 145, 2023
|
|
5G0I
 
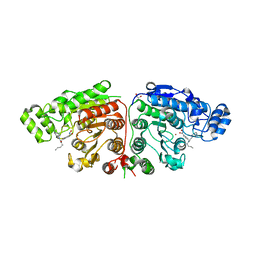 | | Crystal structure of Danio rerio HDAC6 CD1 and CD2 (linker cleaved) in complex with Nexturastat A | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, HDAC6, ... | | 著者 | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | 登録日 | 2016-03-18 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0J
 
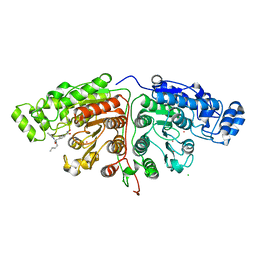 | | Crystal structure of Danio rerio HDAC6 CD1 and CD2 (linker intact) in complex with Nexturastat A | | 分子名称: | CHLORIDE ION, HDAC6, NEXTURASTAT A, ... | | 著者 | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | 登録日 | 2016-03-18 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0G
 
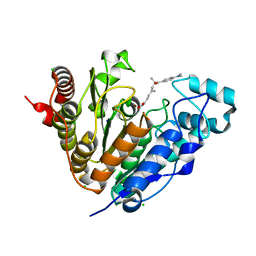 | | Crystal structure of Danio rerio HDAC6 CD1 in complex with trichostatin A | | 分子名称: | CHLORIDE ION, HDAC6, SODIUM ION, ... | | 著者 | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | 登録日 | 2016-03-18 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0F
 
 | | Crystal structure of Danio rerio HDAC6 ZnF-UBP domain | | 分子名称: | GLYCINE, HDAC6, NICKEL (II) ION, ... | | 著者 | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | 登録日 | 2016-03-18 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0H
 
 | | Crystal structure of Danio rerio HDAC6 CD2 in complex with (S)- trichostatin A | | 分子名称: | 1,2-ETHANEDIOL, HDAC6, POTASSIUM ION, ... | | 著者 | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | 登録日 | 2016-03-18 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5YY5
 
 | | Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | 著者 | Zhang, S, Wang, P, Zhou, P, Wang, X, Zhang, L. | | 登録日 | 2017-12-08 | | 公開日 | 2018-08-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Definition of a Unique Neutralization Epitope on the Receptor-Binding Domain of MERS-CoV Spike Glycoprotein
Cell Rep, 24, 2018
|
|
5ZXV
 
 | |
1ZYR
 
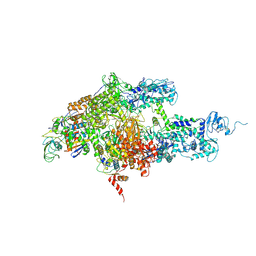 | | Structure of Thermus thermophilus RNA polymerase holoenzyme in complex with the antibiotic streptolydigin | | 分子名称: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase omega chain, ... | | 著者 | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | 登録日 | 2005-06-10 | | 公開日 | 2005-09-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation.
Cell(Cambridge,Mass.), 122, 2005
|
|
2AAB
 
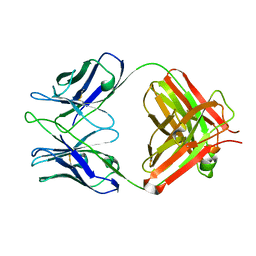 | | Structural basis of antigen mimicry in a clinically relevant melanoma antigen system | | 分子名称: | anti-idiotypic monoclonal antibody (heavy chain), anti-idiotypic monoclonal antibody (light chain) | | 著者 | Chang, C.-C, Hernandez-Guzman, F.G, Luo, W, Wang, X, Ferrone, S, Ghosh, D. | | 登録日 | 2005-07-13 | | 公開日 | 2005-10-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis of antigen mimicry in a clinically relevant melanoma antigen system.
J.Biol.Chem., 280, 2005
|
|
8EY9
 
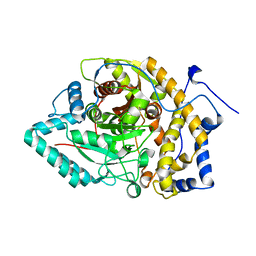 | | Structure of Arabidopsis fatty acid amide hydrolase mutant S305A in complex with 9-hydroxy-10,12-octadecadienoyl-ethanolamide | | 分子名称: | (9R,10E,12Z)-9-hydroxy-N-(2-hydroxyethyl)octadeca-10,12-dienamide, Fatty acid amide hydrolase | | 著者 | Aziz, M, Wang, X, Gaguancela, O.A, Chapman, K.D. | | 登録日 | 2022-10-26 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.59 Å) | | 主引用文献 | Structural interactions explain the versatility of FAAH in the hydrolysis of plant and microbial acyl amide signals
To be published
|
|
8EWW
 
 | |
7RST
 
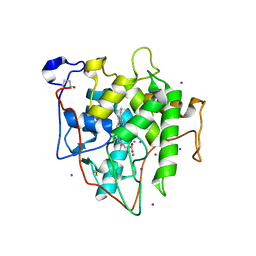 | | The Crystal Structure of Recombinant Chloroperoxidase Expressed in Aspergillus niger | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chloroperoxidase, ... | | 著者 | Tang, X, Venkadesh, S, Zhou, J, Rosen, B, Wang, X. | | 登録日 | 2021-08-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | The Crystal Structure of Recombinant Chloroperoxidase Expressed in Aspergillus niger
To Be Published
|
|
2JS1
 
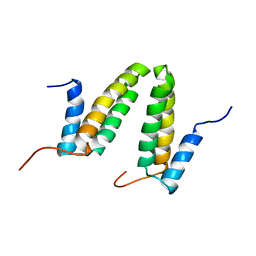 | | Solution NMR structure of the homodimer protein YVFG from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR478 | | 分子名称: | Uncharacterized protein yvfG | | 著者 | Macnaughtan, M.A, Weldeghiorghis, T, Wang, X, Bansal, S, Tian, F, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-06-29 | | 公開日 | 2007-07-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of the Bacillus subtilis Protein YvfG.
To be Published
|
|
8EY1
 
 | |
3V32
 
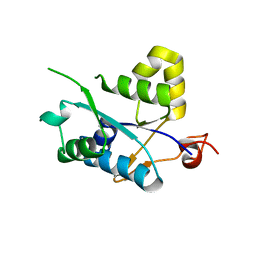 | | Crystal structure of MCPIP1 N-terminal conserved domain | | 分子名称: | Ribonuclease ZC3H12A | | 著者 | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | 登録日 | 2011-12-12 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
3V34
 
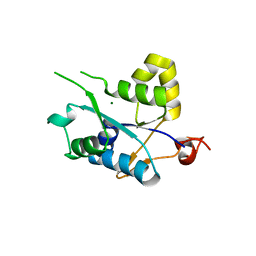 | | Crystal structure of MCPIP1 conserved domain with magnesium ion in the catalytic center | | 分子名称: | MAGNESIUM ION, Ribonuclease ZC3H12A | | 著者 | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | 登録日 | 2011-12-12 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
3V33
 
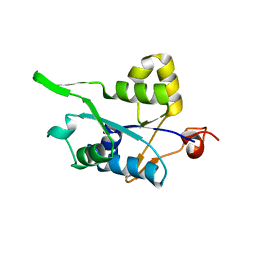 | | Crystal structure of MCPIP1 conserved domain with zinc-finger motif | | 分子名称: | Ribonuclease ZC3H12A | | 著者 | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | 登録日 | 2011-12-12 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.005 Å) | | 主引用文献 | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
5DZN
 
 | |
3SLU
 
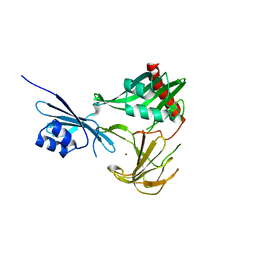 | | Crystal structure of NMB0315 | | 分子名称: | M23 peptidase domain protein, NICKEL (II) ION | | 著者 | Shen, Y, Wang, X, Yang, X, Xu, H. | | 登録日 | 2011-06-26 | | 公開日 | 2012-02-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Crystal structure of outer membrane protein NMB0315 from Neisseria meningitidis.
Plos One, 6, 2011
|
|
