4YGM
 
 | | Vaccinia virus his-D4/A20(1-50) in complex with uracil | | Descriptor: | DNA polymerase processivity factor component A20, SULFATE ION, URACIL, ... | | Authors: | Tarbouriech, N, Iseni, F, Burmeister, W.P. | | Deposit date: | 2015-02-26 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Vaccinia Virus Uracil-DNA Glycosylase in Complex with DNA.
J.Biol.Chem., 290, 2015
|
|
4YIG
 
 | | vaccinia virus D4/A20(1-50) in complex with dsDNA containing an abasic site and free uracyl | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*(ORP)P*AP*TP*CP*TP*T)-3'), DNA polymerase processivity factor component A20, ... | | Authors: | tarbouriech, N, burmeister, W.P, iseni, F. | | Deposit date: | 2015-03-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Vaccinia Virus Uracil-DNA Glycosylase in Complex with DNA.
J.Biol.Chem., 290, 2015
|
|
8QAM
 
 | |
8B8D
 
 | | multimerization domain of Gaboon Viper Virus 1 | | Descriptor: | Phosphoprotein | | Authors: | Tarbouriech, N, Legrand, P, Bouhris, J.M, Horie, M, Tomonaga, K, Crepin, T. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses, 14, 2022
|
|
8B8A
 
 | | Multimerization domain of borna disease virus 1 phosphoprotein | | Descriptor: | Phosphoprotein | | Authors: | Tarbouriech, N, Legrand, P, Bourhis, J.M, Chenavier, F, Freslon, L, Kawasaki, J, Horie, M, Tomonaga, K, Bachiri, K, Coyaud, E, Gonzalez-Dunia, D, Ruigrok, R.W.H, Crepin, T. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses, 14, 2022
|
|
1H7L
 
 | | dTDP-MAGNESIUM COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | MAGNESIUM ION, SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Tarbouriech, N, Charnock, S.J, Davies, G.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Three-Dimensional Structures of the Mn and Mg Dtdp Complexes of the Family Gt-2 Glycosyltransferase Spsa: A Comparison with Related Ndp-Sugar Glycosyltransferases.
J.Mol.Biol., 314, 2001
|
|
1H7Q
 
 | | dTDP-MANGANESE COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA, ... | | Authors: | Tarbouriech, N, Charnock, S.J, Davies, G.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-Dimensional Structures of the Mn and Mg Dtdp Complexes of the Family Gt-2 Glycosyltransferase Spsa: A Comparison with Related Ndp-Sugar Glycosyltransferases.
J.Mol.Biol., 314, 2001
|
|
5N2E
 
 | | Structure of the E9 DNA polymerase from vaccinia virus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tarbouriech, N, Burmeister, W.P, Iseni, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun, 8, 2017
|
|
5N2H
 
 | | Structure of the E9 DNA polymerase exonuclease deficient mutant (D166A+E168A) from vaccinia virus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tarbouriech, N, Burmeister, W.P, Iseni, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun, 8, 2017
|
|
5N2G
 
 | | Structure of the E9 DNA polymerase from vaccinia virus in complex with manganese | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tarbouriech, N, Burmeister, W.P, Iseni, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun, 8, 2017
|
|
1EZJ
 
 | | CRYSTAL STRUCTURE OF THE MULTIMERIZATION DOMAIN OF THE PHOSPHOPROTEIN FROM SENDAI VIRUS | | Descriptor: | CALCIUM ION, ETHYL MERCURY ION, NUCLEOCAPSID PHOSPHOPROTEIN | | Authors: | Tarbouriech, N, Curran, J, Ruigrok, R.W.H, Burmeister, W.P. | | Deposit date: | 2000-05-11 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric coiled coil domain of Sendai virus phosphoprotein.
Nat.Struct.Biol., 7, 2000
|
|
2BSY
 
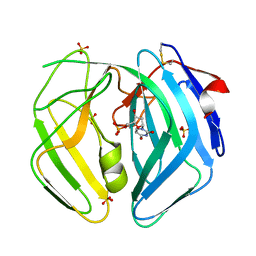 | | Epstein Barr Virus dUTPase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, SULFATE ION | | Authors: | Tarbouriech, N, Buisson, M, Seigneurin, J.-M, Cusack, S, Burmeister, W.P. | | Deposit date: | 2005-05-24 | | Release date: | 2005-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Monomeric Dutpase from Epstein-Barr Virus Mimics Trimeric Dutpases
Structure, 13, 2005
|
|
2BT1
 
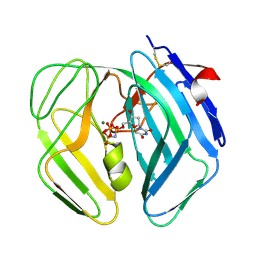 | | Epstein Barr Virus dUTPase in complex with a,b-imino dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, MAGNESIUM ION | | Authors: | Tarbouriech, N, Buisson, M, Seigneurin, J.-M, Cusack, S, Burmeister, W.P. | | Deposit date: | 2005-05-24 | | Release date: | 2005-09-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Monomeric Dutpase from Epstein-Barr Virus Mimics Trimeric Dutpases
Structure, 13, 2005
|
|
2CH8
 
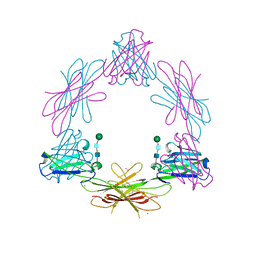 | | Structure of the Epstein-Barr Virus Oncogene BARF1 | | Descriptor: | 33 KDA EARLY PROTEIN, PLATINUM (II) ION, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Tarbouriech, N, Ruggiero, F, deTurenne-Tessier, M, Ooka, T, Burmeister, W.P. | | Deposit date: | 2006-03-13 | | Release date: | 2006-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Epstein-Barr Virus Oncogene Barf1
J.Mol.Biol., 359, 2006
|
|
1WB4
 
 | | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with sinapinate | | Descriptor: | ACETIC ACID, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Tarbouriech, N, Prates, J.A, Fontes, C, Davies, G.J. | | Deposit date: | 2004-10-30 | | Release date: | 2005-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Determinants of Substrate Specificity in the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1WB5
 
 | | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with syringate | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Tarbouriech, N, Prates, J.A, Fontes, C, Davies, G.J. | | Deposit date: | 2004-10-30 | | Release date: | 2005-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Determinants of Substrate Specificity in the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1WB6
 
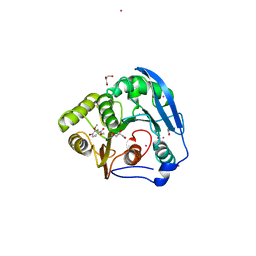 | | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with vanillate | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Tarbouriech, N, Prates, J.A, Fontes, C, Davies, G.J. | | Deposit date: | 2004-10-30 | | Release date: | 2006-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Determinants of Substrate Specificity in the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4WCY
 
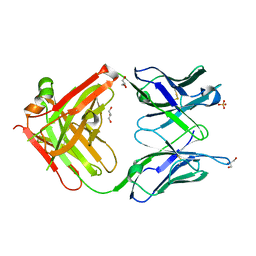 | | Fab fragment of mouse AZ130 monoclonal antibody | | Descriptor: | AZ130 Heavy chain, AZ130 Light chain, GLYCEROL, ... | | Authors: | Fallecker, C, Tarbouriech, N, Habib, M, Crepin, T, Drouet, E. | | Deposit date: | 2014-09-05 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fab fragment of mouse AZ130 monoclonal antibody
To Be Published
|
|
5JKT
 
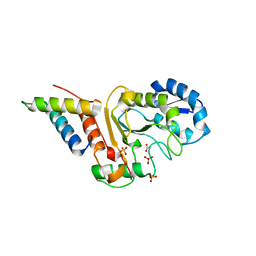 | | vaccinia virus D4 P173G mutant /A20(1-50) | | Descriptor: | ACETATE ION, DNA polymerase processivity factor component A20, SULFATE ION, ... | | Authors: | Contesto-Richefeu, C, Tarbouriech, N, Brazzolotto, X, Burmeister, W.P, Peyrefitte, C.N, Iseni, F. | | Deposit date: | 2016-04-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural analysis of point mutations at the Vaccinia virus A20/D4 interface.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6ZXP
 
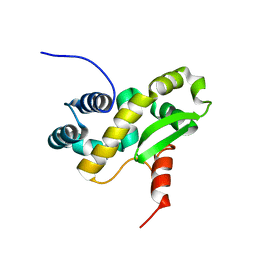 | | Solution structure of the C-terminal domain of the vaccinia virus DNA polymerase processivity factor component A20 fused to a short peptide from the viral DNA polymerase E9. | | Descriptor: | DNA polymerase processivity factor component A20,DNA polymerase processivity factor component E9 | | Authors: | Bersch, B, Tarbouriech, N, Burmeister, W, Iseni, F. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain of A20, the Missing Brick for the Characterization of the Interface between Vaccinia Virus DNA Polymerase and its Processivity Factor.
J.Mol.Biol., 433, 2021
|
|
6ZYC
 
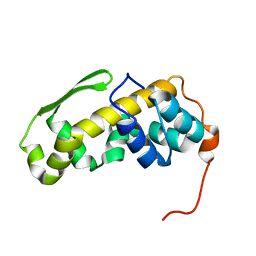 | | Solution structure of the C-terminal domain of the vaccinia virus DNA polymerase processivity factor component A20. | | Descriptor: | DNA polymerase processivity factor component A20 | | Authors: | Bersch, B, Iseni, F, Burmeister, W, Tarbouriech, N. | | Deposit date: | 2020-07-31 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain of A20, the Missing Brick for the Characterization of the Interface between Vaccinia Virus DNA Polymerase and its Processivity Factor.
J.Mol.Biol., 433, 2021
|
|
1R4G
 
 | | Solution structure of the Sendai virus protein X C-subdomain | | Descriptor: | RNA polymerase alpha subunit | | Authors: | Blanchard, L, Tarbouriech, N, Blackledge, M, Timmins, P, Burmeister, W.P, Ruigrok, R.W, Marion, D. | | Deposit date: | 2003-10-06 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of the nucleocapsid-binding domain of the Sendai virus phosphoprotein in solution
Virology, 319, 2004
|
|
7PON
 
 | |
2J8X
 
 | | Epstein-Barr virus uracil-DNA glycosylase in complex with Ugi from PBS-2 | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR, UREA | | Authors: | Geoui, T, Buisson, M, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2006-10-31 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New Insights on the Role of the Gamma-Herpesvirus Uracil-DNA Glycosylase Leucine Loop Revealed by the Structure of the Epstein-Barr Virus Enzyme in Complex with an Inhibitor Protein.
J.Mol.Biol., 366, 2007
|
|
7PNO
 
 | | C terminal domain of Nipah Virus Phosphoprotein fused to the Ntail alpha more of the Nucleoprotein. | | Descriptor: | Phosphoprotein, alpha MoRE of Nipah virus Nucleoprotein tail | | Authors: | Bourhis, J.M, Yabukaski, F, Tarbouriech, N, Jamin, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Dynamics of the C-terminal X Domain of Nipah and Hendra Viruses Controls the Attachment to the C-terminal Tail of the Nucleocapsid Protein.
J.Mol.Biol., 434, 2022
|
|
