3M8F
 
 | |
3JS6
 
 | |
3MKW
 
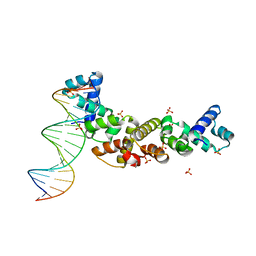 | | Structure of sopB(155-272)-18mer complex, I23 form | | 分子名称: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | 著者 | Schumacher, M.A, Piro, K, Xu, W. | | 登録日 | 2010-04-15 | | 公開日 | 2010-05-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3M9A
 
 | |
4DZZ
 
 | | Structure of ParF-ADP, crystal form 1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid partitioning protein ParF | | 著者 | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | 登録日 | 2012-03-01 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4E09
 
 | | Structure of ParF-AMPPCP, I422 form | | 分子名称: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Plasmid partitioning protein ParF, SULFATE ION | | 著者 | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | 登録日 | 2012-03-02 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4E07
 
 | | ParF-AMPPCP-C2221 form | | 分子名称: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Plasmid partitioning protein ParF | | 著者 | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | 登録日 | 2012-03-02 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
3MKY
 
 | | Structure of SopB(155-323)-18mer DNA complex, I23 form | | 分子名称: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | 著者 | Schumacher, M.A, Piro, K, Xu, W. | | 登録日 | 2010-04-15 | | 公開日 | 2010-05-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
4GFK
 
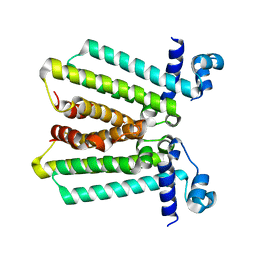 | | structures of NO factors | | 分子名称: | Nucleoid occlusion factor SlmA | | 著者 | Schumacher, M.A. | | 登録日 | 2012-08-03 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7TDP
 
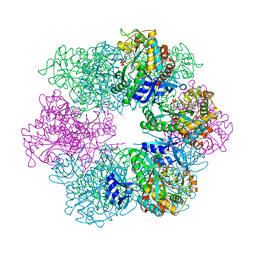 | |
7U02
 
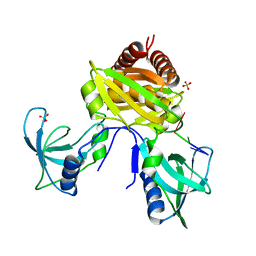 | |
7TEA
 
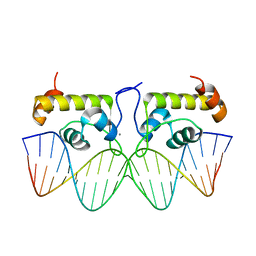 | | Crystal structure of S. aureus GlnR-DNA complex | | 分子名称: | CALCIUM ION, DNA (5'-D(*CP*GP*TP*GP*TP*CP*AP*GP*AP*TP*AP*AP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*GP*TP*CP*AP*GP*AP*TP*TP*AP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), ... | | 著者 | Schumacher, M.A. | | 登録日 | 2022-01-04 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TEC
 
 | |
7TDV
 
 | |
7TEN
 
 | |
7U3A
 
 | | Structure of the Streptomyces venezuelae GlgX-c-di-GMP complex | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX | | 著者 | Schumacher, M.A. | | 登録日 | 2022-02-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.34 Å) | | 主引用文献 | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
7U3D
 
 | | Structure of S. venezuelae GlgX-c-di-GMP-acarbose complex (4.6) | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX | | 著者 | Schumacher, M.A. | | 登録日 | 2022-02-27 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
4YG4
 
 | | HipB-O1-O1* complex | | 分子名称: | Antitoxin HipB, DNA (28-MER), DNA (5'-D(*AP*TP*AP*TP*CP*CP*CP*CP*TP*TP*AP*AP*GP*GP*GP*GP*AP*TP*AP*A)-3') | | 著者 | Schumacher, M.A. | | 登録日 | 2015-02-25 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | HipBA-promoter structures reveal the basis of heritable multidrug tolerance.
Nature, 524, 2015
|
|
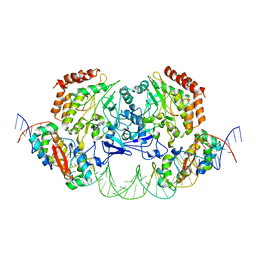
4YG7
 
 | |
7TZV
 
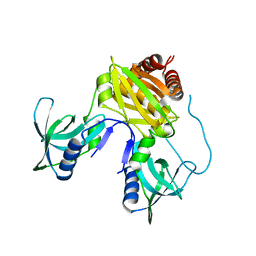 | | Structure of DriD C-domain bound to 9mer ssDNA | | 分子名称: | DNA (5'-D(*TP*AP*GP*TP*CP*TP*AP*CP*T)-3'), WYL domain-containing protein | | 著者 | Schumacher, M.A, Laub, M. | | 登録日 | 2022-02-16 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | ssDNA is an allosteric regulator of the C. crescentus SOS-independent DNA damage response transcription activator, DriD.
Genes Dev., 36, 2022
|
|
7U3B
 
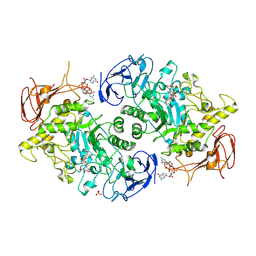 | | Structure of S. venezuelae GlgX bound to c-di-GMP and acarbose (pH 8.5) | | 分子名称: | 4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranosyl)-beta-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX, ... | | 著者 | Schumacher, M.A, Tschowri, N. | | 登録日 | 2022-02-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
7U39
 
 | |
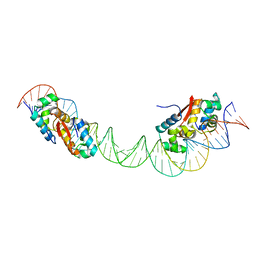
4YG1
 
 | |
3VEA
 
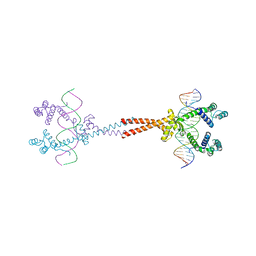 | | Crystal Structure of matP-matS23mer | | 分子名称: | 5'-D(*AP*GP*TP*TP*CP*GP*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*CP*GP*AP*AP*CP*T)-3', 5'-D(*AP*GP*TP*TP*CP*GP*TP*GP*AP*CP*AP*TP*TP*GP*TP*CP*AP*CP*GP*AP*AP*CP*T)-3', Macrodomain Ter protein | | 著者 | Schumacher, M.A. | | 登録日 | 2012-01-07 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome.
Mol.Cell, 48, 2012
|
|
3VEB
 
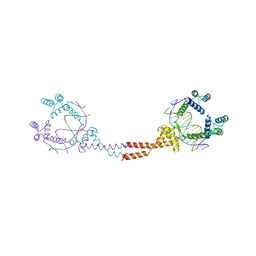 | | Crystal Structure of Matp-matS | | 分子名称: | 5'-D(*AP*CP*GP*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*CP*G)-3', 5'-D(*TP*CP*GP*TP*GP*AP*CP*AP*TP*TP*GP*TP*CP*AP*CP*G)-3', CALCIUM ION, ... | | 著者 | Schumacher, M.A. | | 登録日 | 2012-01-07 | | 公開日 | 2012-11-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome.
Mol.Cell, 48, 2012
|
|
