4PTA
 
 | | Structure of MDR initiator | | Descriptor: | Replication initiator protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6003 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PQK
 
 | | C-Terminal domain of DNA binding protein | | Descriptor: | Maltose ABC transporter periplasmic protein, Truncated replication protein RepA, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N.K. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3TPE
 
 | | The phipa p3121 structure | | Descriptor: | Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A, Link, T, Brennan, R.G. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPV
 
 | | Structure of pHipA bound to ADP | | Descriptor: | ADENINE, SULFATE ION, Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A, Link, T.M, Brennan, R.G. | | Deposit date: | 2011-09-08 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPT
 
 | | Structure of HipA(D309Q) bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | schumacher, M.A, link, T, Brennan, R.G. | | Deposit date: | 2011-09-08 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
4OB4
 
 | | Structure of the S. venezulae BldD DNA-binding domain | | Descriptor: | Putative DNA-binding protein | | Authors: | schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
4OAY
 
 | | BldD CTD-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R.G. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
3TPD
 
 | | Structure of pHipA, monoclinic form | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Serine/threonine-protein kinase HipA | | Authors: | schumacher, M.A, link, T, Brennan, R.G. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
4OAZ
 
 | | BldD CTD-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative DNA-binding protein | | Authors: | Schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R.G. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
3TPB
 
 | | Structure of HipA(S150A) | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Serine/threonine-protein kinase HipA | | Authors: | schumacher, M.A. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3KZ5
 
 | | Structure of cdomain | | Descriptor: | ACETATE ION, Protein sopB | | Authors: | Schumacher, M.A. | | Deposit date: | 2009-12-07 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3M8K
 
 | | Protein structure of type III plasmid segregation TubZ | | Descriptor: | FtsZ/tubulin-related protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2010-03-18 | | Release date: | 2010-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MKZ
 
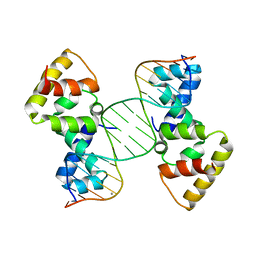 | | Structure of SopB(155-272)-18mer complex, P21 form | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB | | Authors: | Schumacher, M.A. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3M8F
 
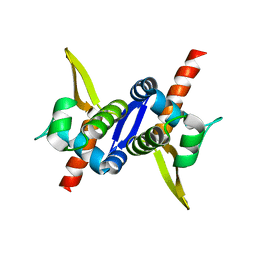 | | Protein structure of type III plasmid segregation TubR mutant | | Descriptor: | Putative DNA-binding protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JS6
 
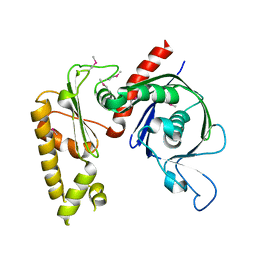 | | Crystal structure of apo psk41 parM protein | | Descriptor: | Uncharacterized ParM protein | | Authors: | Schumacher, M.A, Xu, W, Firth, N. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation.
J.Biol.Chem., 285, 2010
|
|
3MKW
 
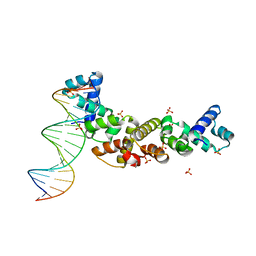 | | Structure of sopB(155-272)-18mer complex, I23 form | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | Authors: | Schumacher, M.A, Piro, K, Xu, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3M9A
 
 | | Protein structure of type III plasmid segregation TubR | | Descriptor: | Putative DNA-binding protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2010-03-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MKY
 
 | | Structure of SopB(155-323)-18mer DNA complex, I23 form | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | Authors: | Schumacher, M.A, Piro, K, Xu, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3OQM
 
 | | structure of ccpa-hpr-ser46p-ackA2 complex | | Descriptor: | 5'-D(*TP*TP*GP*AP*TP*AP*AP*CP*GP*CP*TP*TP*AP*CP*AP*A)-3', 5'-D(*TP*TP*GP*TP*AP*AP*GP*CP*GP*TP*TP*AP*TP*CP*AP*A)-3', Catabolite control protein A, ... | | Authors: | Schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
3Q5W
 
 | | Structure of proteasome tether | | Descriptor: | Protein cut8 | | Authors: | Schumacher, M.A. | | Deposit date: | 2010-12-30 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Proteasome Tether
To be Published
|
|
3PM1
 
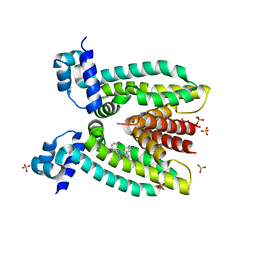 | | Structure of QacR E90Q bound to Ethidium | | Descriptor: | ETHIDIUM, HTH-type transcriptional regulator qacR, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A single acidic residue can guide binding site selection but does not govern QacR cationic-drug affinity.
Plos One, 6, 2011
|
|
3OQN
 
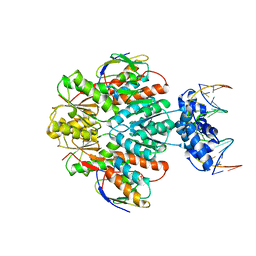 | | Structure of ccpa-hpr-ser46-p-gntr-down cre | | Descriptor: | 5'-D(*AP*TP*GP*GP*TP*AP*CP*CP*GP*CP*TP*TP*TP*CP*AP*A)-3', 5'-D(*TP*TP*GP*AP*AP*AP*GP*CP*GP*GP*TP*AP*CP*CP*AP*T)-3', Catabolite control protein A, ... | | Authors: | Schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
3OQO
 
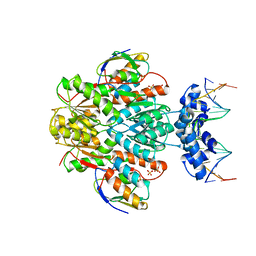 | | Ccpa-hpr-ser46p-syn cre | | Descriptor: | 5'-D(*CP*TP*GP*AP*AP*AP*GP*CP*GP*CP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*AP*GP*CP*GP*CP*TP*TP*TP*CP*AP*G)-3', Catabolite control protein A, ... | | Authors: | schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
3Q5X
 
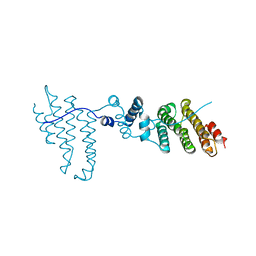 | | Structure of proteasome tether | | Descriptor: | Protein cut8 | | Authors: | Schumacher, M.A. | | Deposit date: | 2010-12-30 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure of Proteasome Tether
To be Published
|
|
3VEA
 
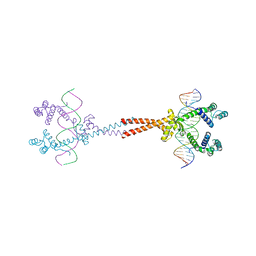 | | Crystal Structure of matP-matS23mer | | Descriptor: | 5'-D(*AP*GP*TP*TP*CP*GP*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*CP*GP*AP*AP*CP*T)-3', 5'-D(*AP*GP*TP*TP*CP*GP*TP*GP*AP*CP*AP*TP*TP*GP*TP*CP*AP*CP*GP*AP*AP*CP*T)-3', Macrodomain Ter protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-01-07 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome.
Mol.Cell, 48, 2012
|
|
