8WDN
 
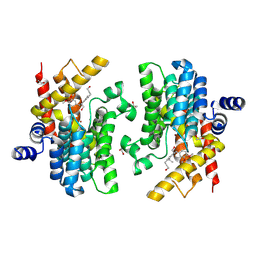 | | Crystal structure of PDE4D complexed with 7b-1 | | 分子名称: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-7-ethoxy-2-(3-ethoxy-4-methoxy-phenyl)-3-(hydroxymethyl)-2,3-dihydro-1-benzofuran-5-yl]propan-1-ol, MAGNESIUM ION, ... | | 著者 | Liu, J.Y, Li, M.J, Xu, Y.C. | | 登録日 | 2023-09-15 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Identification of Dihydrobenzofuran Neolignans as Novel PDE4 Inhibitors and Evaluation of Antiatopic Dermatitis Efficacy in DNCB-Induced Mice Model.
J.Med.Chem., 67, 2024
|
|
9BA4
 
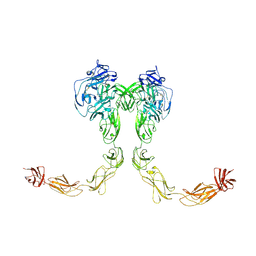 | | Full-length cross-linked Contactin 2 (CNTN2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-2 | | 著者 | Liu, J.L, Fan, S.F, Ren, G.R, Rudenko, G.R. | | 登録日 | 2024-04-03 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Molecular mechanism of contactin 2 homophilic interaction.
Structure, 2024
|
|
9BA5
 
 | |
6V9C
 
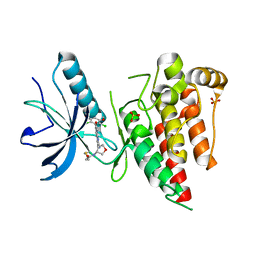 | | Crystal structure of FGFR4 kinase domain in complex with covalent inhibitor | | 分子名称: | Fibroblast growth factor receptor 4, N-[(3R,4S)-4-{[6-(2,6-dichloro-3,5-dimethoxyphenyl)-8-methyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-2-yl]amino}oxolan-3-yl]prop-2-enamide, SULFATE ION | | 著者 | Liu, J, Liu, H. | | 登録日 | 2019-12-13 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of Selective, Covalent FGFR4 Inhibitors with Antitumor Activity in Models of Hepatocellular Carcinoma.
Acs Med.Chem.Lett., 11, 2020
|
|
7AX3
 
 | |
6ZVR
 
 | | C11 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | 分子名称: | Vipp1 | | 著者 | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | 登録日 | 2020-07-27 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW4
 
 | | C14 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | 分子名称: | vipp1 | | 著者 | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | 登録日 | 2020-07-27 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW5
 
 | | C15 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | 分子名称: | vipp1 | | 著者 | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | 登録日 | 2020-07-27 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZVT
 
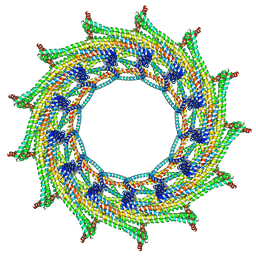 | | C13 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | 分子名称: | Vipp1 | | 著者 | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | 登録日 | 2020-07-27 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6NY1
 
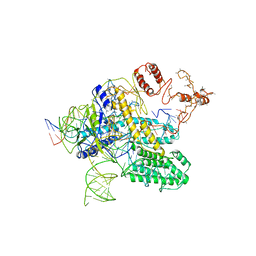 | | CasX-gRNA-DNA(30bp) State II | | 分子名称: | CasX, DNA Non-target strand, DNA target strand, ... | | 著者 | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | 登録日 | 2019-02-10 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
3ZP3
 
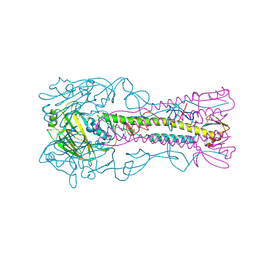 | | INFLUENZA VIRUS (VN1194) H5 HA A138V mutant with LSTc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ, N-acetyl-alpha-neuraminic acid, ... | | 著者 | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | 登録日 | 2013-02-26 | | 公開日 | 2013-10-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
3ZP1
 
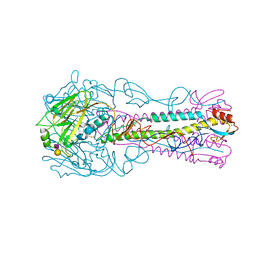 | | INFLUENZA VIRUS (VN1194) H5 HA with LSTc | | 分子名称: | HAEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-2)-beta-D-galactopyranose | | 著者 | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | 登録日 | 2013-02-26 | | 公開日 | 2013-10-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
3ZP2
 
 | | INFLUENZA VIRUS (VN1194) H5 HA A138V mutant with LSTa | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose | | 著者 | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | 登録日 | 2013-02-26 | | 公開日 | 2013-10-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
3ZP6
 
 | | INFLUENZA VIRUS (VN1194) H5 E190D mutant HA with LSTc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose | | 著者 | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | 登録日 | 2013-02-26 | | 公開日 | 2013-10-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
3ZP0
 
 | | INFLUENZA VIRUS (VN1194) H5 HA with LSTa | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose | | 著者 | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | 登録日 | 2013-02-26 | | 公開日 | 2013-10-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
5OXF
 
 | |
4YJ5
 
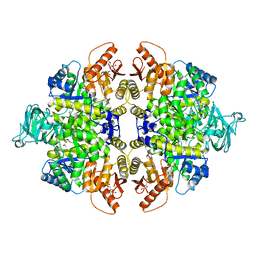 | | Crystal structure of PKM2 mutant | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PYRUVIC ACID, ... | | 著者 | Liu, J.-S, Wu, C.-W, Wang, W.-C. | | 登録日 | 2015-03-03 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Mutations in the PKM2 exon-10 region are associated with reduced allostery and increased nuclear translocation
Commun Biol, 2, 2019
|
|
3ZPA
 
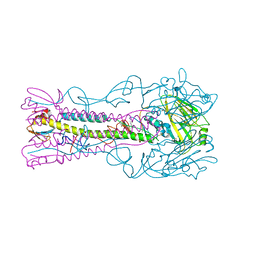 | | INFLUENZA VIRUS (VN1194) H5 I155F mutant HA | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ | | 著者 | Liu, J, Chen, Z, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | 登録日 | 2013-02-27 | | 公開日 | 2013-10-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
3ZPB
 
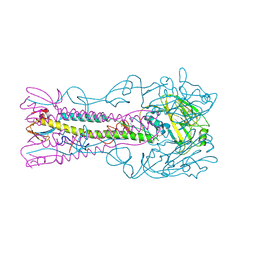 | | INFLUENZA VIRUS (VN1194) H5 E190D mutant HA with LSTa | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | 登録日 | 2013-02-27 | | 公開日 | 2013-10-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
5G06
 
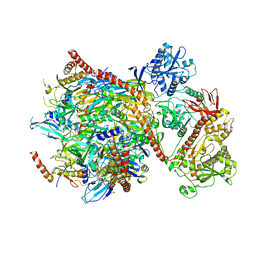 | | Cryo-EM structure of yeast cytoplasmic exosome | | 分子名称: | EXOSOME COMPLEX COMPONENT CSL4, EXOSOME COMPLEX COMPONENT MTR3, EXOSOME COMPLEX COMPONENT RRP4, ... | | 著者 | Liu, J.J, Niu, C.Y, Wu, Y, Tan, D, Wang, Y, Ye, M.D, Liu, Y, Zhao, W.W, Zhou, K, Liu, Q.S, Dai, J.B, Yang, X.R, Dong, M.Q, Huang, N, Wang, H.W. | | 登録日 | 2016-03-17 | | 公開日 | 2016-06-15 | | 最終更新日 | 2017-08-02 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryoem Structure of Yeast Cytoplasmic Exosome Complex.
Cell Res., 26, 2016
|
|
5OWV
 
 | |
8PE9
 
 | | Complex between DDR1 DS-like domain and PRTH-101 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Liu, J, Chiang, H, Xiong, W, Laurent, V, Griffiths, S.C, Duelfer, J, Deng, H, Sun, X, Yin, Y.W, Li, W, Audoly, L.P, An, Z, Schuerpf, T, Li, R, Zhang, N. | | 登録日 | 2023-06-13 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.152 Å) | | 主引用文献 | A highly selective humanized DDR1 mAb reverses immune exclusion by disrupting collagen fiber alignment in breast cancer.
J Immunother Cancer, 11, 2023
|
|
6ZVS
 
 | | C12 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | 分子名称: | Vipp1 | | 著者 | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | 登録日 | 2020-07-27 | | 公開日 | 2021-08-04 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW6
 
 | | C16 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | 分子名称: | vipp1 | | 著者 | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | 登録日 | 2020-07-27 | | 公開日 | 2021-08-04 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (7.4 Å) | | 主引用文献 | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW7
 
 | | C17 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | 分子名称: | vipp1 | | 著者 | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | 登録日 | 2020-07-27 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (9.4 Å) | | 主引用文献 | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
