7CN3
 
 | | 2,5-dihydroxypridine Dioxygenase in complex with 2,5-dihydroxypridine and product N-formylmaleamic acid | | Descriptor: | (~{Z})-4-formamido-4-oxidanylidene-but-2-enoic acid, 1,2-ETHANEDIOL, 2,5-dihydroxypyridine 5,6-dioxygenase, ... | | Authors: | Liu, G.Q, Tang, H.Z. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2,5-dihydroxypridine Dioxygenase in complex with 2,5-dihydroxypridine and product N-formylmaleamic acid
Nat Commun, 2020
|
|
5N9Y
 
 | | The full-length structure of ZntB | | Descriptor: | Zinc transport protein ZntB | | Authors: | Cornelius, G, Stetsenko, A, Scheres, S.H.W, Slotboom, D.J, Guskov, A. | | Deposit date: | 2017-02-27 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis of proton driven zinc transport by ZntB.
Nat Commun, 8, 2017
|
|
1CEE
 
 | | SOLUTION STRUCTURE OF CDC42 IN COMPLEX WITH THE GTPASE BINDING DOMAIN OF WASP | | Descriptor: | GTP-BINDING RHO-LIKE PROTEIN, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Abdul-Manan, N, Aghazadeh, B, Liu, G.A, Majumdar, A, Ouerfelli, O, Rosen, M.K. | | Deposit date: | 1999-03-08 | | Release date: | 1999-06-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Cdc42 in complex with the GTPase-binding domain of the 'Wiskott-Aldrich syndrome' protein.
Nature, 399, 1999
|
|
2DO8
 
 | | Solution Structure of UPF0301 protein HD_1794 | | Descriptor: | UPF0301 protein HD_1794 | | Authors: | Zhang, Q, Liu, G, Shastry, R, Jiang, M, Cunningham, K, Ma, L.C, Xiao, R, Acton, T.R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-27 | | Release date: | 2006-05-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of UPF0301 protein HD_1794
To be published
|
|
1KLP
 
 | | The Solution Structure of Acyl Carrier Protein from Mycobacterium tuberculosis | | Descriptor: | MEROMYCOLATE EXTENSION ACYL CARRIER PROTEIN | | Authors: | Wong, H.C, Liu, G, Zhang, Y.-M, Rock, C.O, Zheng, J. | | Deposit date: | 2001-12-12 | | Release date: | 2002-06-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of acyl carrier protein from Mycobacterium tuberculosis.
J.Biol.Chem., 277, 2002
|
|
6E4H
 
 | | Solution NMR Structure of the Colied-coil PALB2 Homodimer | | Descriptor: | Partner and localizer of BRCA2 | | Authors: | Song, F, Li, M, Liu, G, Swapna, G.V.T, Xia, B, Bunting, S.F, Montelione, G.T. | | Deposit date: | 2018-07-17 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antiparallel Coiled-Coil Interactions Mediate the Homodimerization of the DNA Damage-Repair Protein PALB2.
Biochemistry, 57, 2018
|
|
6E4J
 
 | | Solution NMR Structure of Protein PF2048.1 | | Descriptor: | Uncharacterized protein PF2048.1 | | Authors: | Daigham, N.S, Liu, G, Swapna, G.V.T, Cole, C, Valafar, H, Montelione, G.T. | | Deposit date: | 2018-07-17 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | REDCRAFT: A Computational Platform Using Residual Dipolar Coupling NMR Data for Determining Structures of Perdeuterated Proteins Without NOEs
To Be Published
|
|
1EJ5
 
 | | SOLUTION STRUCTURE OF THE AUTOINHIBITED CONFORMATION OF WASP | | Descriptor: | WISKOTT-ALDRICH SYNDROME PROTEIN | | Authors: | Kim, A.S, Kakalis, L.T, Abdul-Manan, N, Liu, G.A, Rosen, M.K. | | Deposit date: | 2000-02-29 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein.
Nature, 404, 2000
|
|
1FI7
 
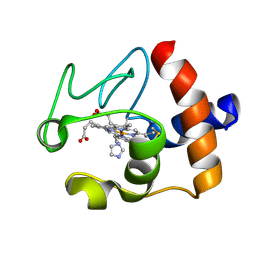 | | Solution structure of the imidazole complex of cytochrome C | | Descriptor: | CYTOCHROME C, HEME C, IMIDAZOLE | | Authors: | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
1PYN
 
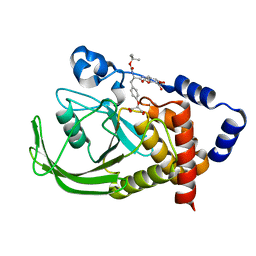 | | DUAL-SITE POTENT, SELECTIVE PROTEIN TYROSINE PHOSPHATASE 1B INHIBITOR USING A LINKED FRAGMENT STRATEGY AND A MALONATE HEAD ON THE FIRST SITE | | Descriptor: | 2-(4-{2-TERT-BUTOXYCARBONYLAMINO-2-[4-(3-HYDROXY-2-METHOXYCARBONYL-PHENOXY)-BUTYLCARBAMOYL]-ETHYL}-PHENOXY)-MALONIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Zhonghua, P, Lubben, T, Trevillyan, J.M, Stashko, M, Ballaron, S.J, Liang, H. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and SAR of novel, potent and selective protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
3J17
 
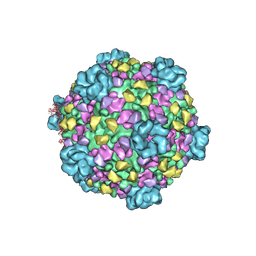 | | Structure of a transcribing cypovirus by cryo-electron microscopy | | Descriptor: | Structural protein VP3, Structural protein VP5, VP1 | | Authors: | Yang, C, Ji, G, Liu, H, Zhang, K, Liu, G, Sun, F, Zhu, P, Cheng, L. | | Deposit date: | 2011-12-25 | | Release date: | 2012-04-04 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a transcribing cypovirus.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3Q9U
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | Descriptor: | COENZYME A, CoA binding protein, consensus ankyrin repeat | | Authors: | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-01-10 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
3Q9N
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | Descriptor: | CARBAMOYL SARCOSINE, COENZYME A, CoA binding protein, ... | | Authors: | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-01-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
5KPH
 
 | |
5KPE
 
 | | Solution NMR Structure of Denovo Beta Sheet Design Protein, Northeast Structural Genomics Consortium (NESG) Target OR664 | | Descriptor: | De novo Beta Sheet Design Protein OR664 | | Authors: | Tang, Y, Liu, G, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-07-03 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5D8W
 
 | | Structrue of a lucidum protein | | Descriptor: | Endoglucanase | | Authors: | Guo, R, Li, Q, Shang, N, Liu, G, Ko, T.P, Chen, C.C, Liu, W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Functional and structural analyses of a 1,4-beta-endoglucanase from Ganoderma lucidum.
Enzyme.Microb.Technol., 86, 2016
|
|
5D8Z
 
 | | Structrue of a lucidum protein | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, endoglucanase | | Authors: | Guo, R, Li, Q, Shang, N, Liu, G, Ko, T.P, Chen, C.C, Liu, W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional and structural analyses of a 1,4-beta-endoglucanase from Ganoderma lucidum.
Enzyme.Microb.Technol., 86, 2016
|
|
7K3S
 
 | |
8YN7
 
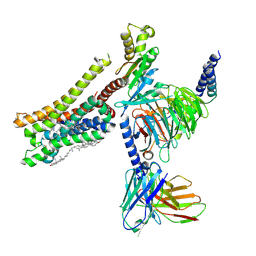 | | Cryo-EM structure of histamine H3 receptor in complex with immethridine and miniGo | | Descriptor: | 4-(1H-imidazol-4-ylmethyl)pyridine, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN3
 
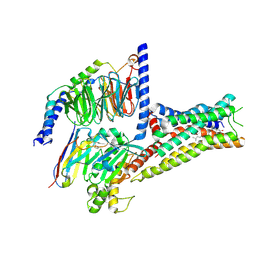 | | Cryo-EM structure of histamine H2 receptor in complex with histamine and miniGs | | Descriptor: | CHOLESTEROL, Engineered guanine nucleotide,binding protein G(s) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN8
 
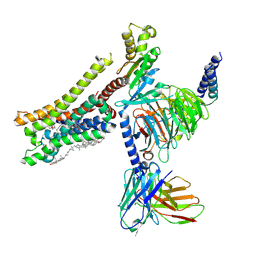 | | Cryo-EM structure of histamine H3 receptor in complex with proxyfan and miniGo | | Descriptor: | 5-(3-phenylmethoxypropyl)-1H-imidazole, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YNA
 
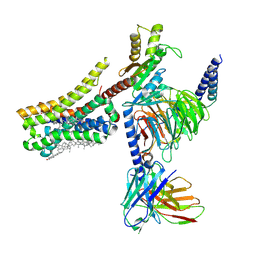 | | Cryo-EM structure of histamine H4 receptor in complex with immepip and Gi | | Descriptor: | 4-(1H-imidazol-5-ylmethyl)piperidine, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN9
 
 | | Cryo-EM structure of histamine H4 receptor in complex with histamine and Gi | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN5
 
 | | Cryo-EM structure of histamine H3 receptor in complex with histamine and Gi | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN2
 
 | | Cryo-EM structure of histamine H1 receptor in complex with histamine and miniGq | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Engineered guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
