7E1Y
 
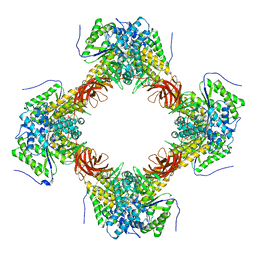 | |
5ZL4
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 wihout its lid in complex with GF2 | | Descriptor: | DFA-IIIase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZL5
 
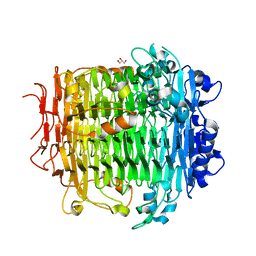 | | Crystal structure of DFA-IIIase mutant C387A from Arthrobacter chlorophenolicus A6 | | Descriptor: | DFA-IIIase C387A mutant, GLYCEROL | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKU
 
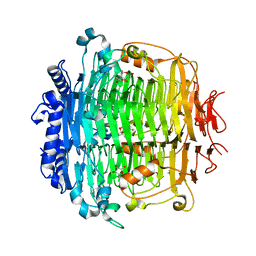 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with DFA-III | | Descriptor: | (2R,3'S,4'S,4aR,5'R,6R,7R,7aS)-4a,5',6-tris(hydroxymethyl)spiro[3,6,7,7a-tetrahydrofuro[2,3-b][1,4]dioxine-2,2'-oxolane ]-3',4',7-triol, DFA-IIIase | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKY
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 without its lid | | Descriptor: | DFA-IIIase | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKS
 
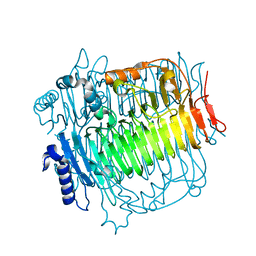 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 | | Descriptor: | DFA-IIIase | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
4IF2
 
 | |
5H5Z
 
 | | Crystal structure of bony fish MHC class I, peptide and B2m II | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide chain | | Authors: | Chen, Z, Zhang, N, Qi, J, Li, X, Chen, R, Wang, Z, Gao, F.G, Xia, C. | | Deposit date: | 2016-11-10 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Mechanism of beta 2m Molecule-Induced Changes in the Peptide Presentation Profile in a Bony Fish.
Iscience, 23, 2020
|
|
7XEE
 
 | | Crystal Structure of the Y53F/N55A mutant of LEH complexed with 2-(3-phenyloxetan-3-yl)ethanamine | | Descriptor: | 1,2-ETHANEDIOL, 2-(3-phenyloxetan-3-yl)ethanamine, Limonene-1,2-epoxide hydrolase, ... | | Authors: | Qu, G, Li, X, Sun, Z.T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7XEF
 
 | | Crystal Structure of the Y53F/N55A mutant of LEH complexed with (R)-(1-benzyl-3-phenylpyrrolidin-3-yl)methanol | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Limonene-1,2-epoxide hydrolase, ... | | Authors: | Qu, G, Li, X, Sun, Z.T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7EXT
 
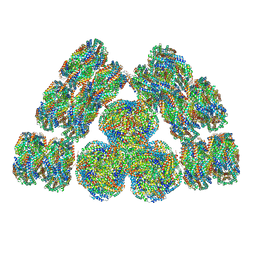 | | Cryo-EM structure of cyanobacterial phycobilisome from Synechococcus sp. PCC 7002 | | Descriptor: | Allophycocyanin alpha subunit, Allophycocyanin beta subunit, Allophycocyanin subunit alpha-B, ... | | Authors: | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | Deposit date: | 2021-05-28 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
7EYD
 
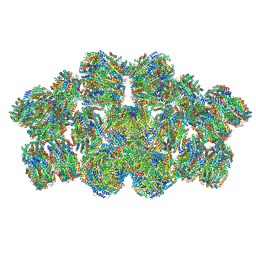 | | Cryo-EM structure of cyanobacterial phycobilisome from Anabaena sp. PCC 7120 | | Descriptor: | Allophycocyanin subunit alpha 1, Allophycocyanin subunit alpha-B, Allophycocyanin subunit beta, ... | | Authors: | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | Deposit date: | 2021-05-30 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
7XL5
 
 | | Crystal structure of the H42T/A85G/I86A mutant of a nadp-dependent alcohol dehydrogenase | | Descriptor: | NADP-dependent isopropanol dehydrogenase | | Authors: | Jiang, Y.Y, Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Engineering the hydrogen transfer pathway of an alcohol dehydrogenase to increase activity by rational enzyme design
Mol Catal, 530, 2022
|
|
8JBZ
 
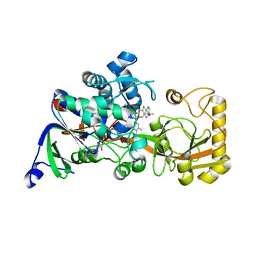 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 4-androstadiene-3,17- dione | | Descriptor: | 3-ketosteroid dehydrogenase, 4-ANDROSTENE-3-17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-05-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 4-androstadiene-3,17- dione
To Be Published
|
|
7W7F
 
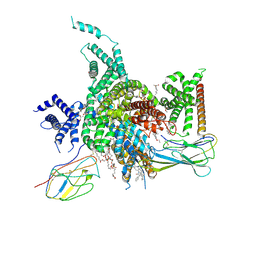 | | Cryo-EM structure of human NaV1.3/beta1/beta2-ICA121431 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2,2-diphenyl-~{N}-[4-(1,3-thiazol-2-ylsulfamoyl)phenyl]ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Li, X. | | Deposit date: | 2021-12-04 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis for modulation of human Na V 1.3 by clinical drug and selective antagonist.
Nat Commun, 13, 2022
|
|
7W77
 
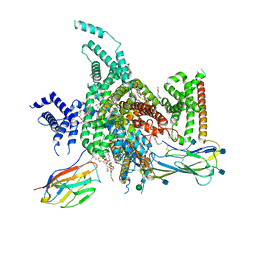 | | cryo-EM structure of human NaV1.3/beta1/beta2-bulleyaconitineA | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Li, X. | | Deposit date: | 2021-12-03 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for modulation of human Na V 1.3 by clinical drug and selective antagonist.
Nat Commun, 13, 2022
|
|
7D8G
 
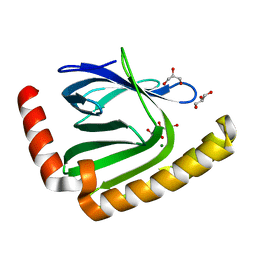 | |
7D8L
 
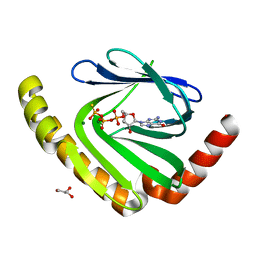 | |
7D8I
 
 | |
7D8Q
 
 | | The structure of nucleotide phosphatase Sa1684 complex with GDP analogue from Staphylococcus aureus | | Descriptor: | MAGNESIUM ION, UPF0374 protein SAB1800c, [(2R,3R,4S,5S)-5-(2-azanyl-6-oxidanyl-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl bis(oxidanyl)phosphinothioyl hydrogen phosphate | | Authors: | Wang, Z, Li, X. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural mechanism for the nucleoside tri- and diphosphate hydrolysis activity of Ntdp from Staphylococcus aureus.
Febs J., 288, 2021
|
|
7DLY
 
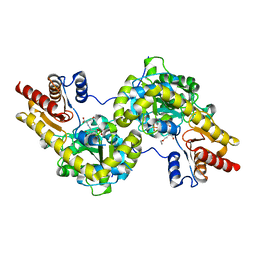 | | Crystal structure of Arabidopsis ACS7 mutant in complex with PPG | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, 1-aminocyclopropane-1-carboxylate synthase 7 | | Authors: | Hao, B, Zhang, Y, Li, X, Rao, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Dual activities of ACC synthase: Novel clues regarding the molecular evolution of ACS genes.
Sci Adv, 7, 2021
|
|
7DLW
 
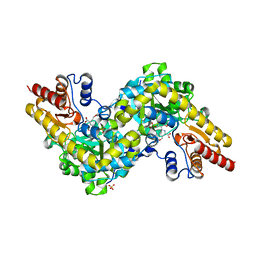 | | Crystal structure of Arabidopsis ACS7 in complex with PPG | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, 1-aminocyclopropane-1-carboxylate synthase 7, SULFATE ION | | Authors: | Hao, B, Zhang, Y, Li, X, Rao, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Dual activities of ACC synthase: Novel clues regarding the molecular evolution of ACS genes.
Sci Adv, 7, 2021
|
|
7DVL
 
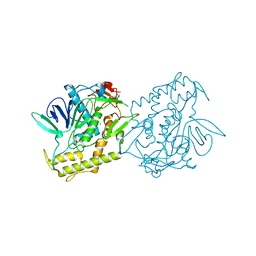 | | Crystal Structure of the Catalytic Domain of Botulinum Neurotoxin Subtype A3 | | Descriptor: | Bont/A3, ZINC ION | | Authors: | Wu, Y, Leka, O, Kammerer, R, Li, X. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of the catalytic domain of botulinum neurotoxin subtype A3.
J.Biol.Chem., 296, 2021
|
|
7ENH
 
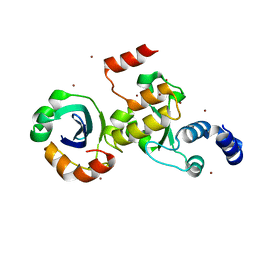 | | Crystal structure of cas and anti-cas protein complex | | Descriptor: | AcrIIA14 protein, CRISPR-associated endonuclease Cas9, NICKEL (II) ION | | Authors: | Wang, Y, Li, X. | | Deposit date: | 2021-04-17 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of cas and anti-cas protein complex
To Be Published
|
|
7ENI
 
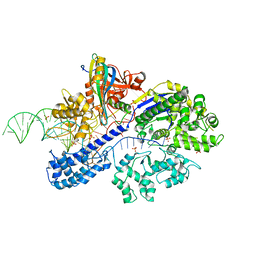 | | Crystal structure of cas and anti-cas protein complex | | Descriptor: | AcrIIA13 protein, CRISPR-associated endonuclease Cas9, PHOSPHATE ION, ... | | Authors: | Wang, Y, Li, X. | | Deposit date: | 2021-04-17 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.632 Å) | | Cite: | Crystal structure of cas and anti-cas protein complex
To Be Published
|
|
