1PJ7
 
 | | Structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with folinic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N,N-dimethylglycine oxidase, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Leys, D, Basran, J, Scrutton, N.S. | | Deposit date: | 2003-06-01 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
Embo J., 22, 2003
|
|
5LW6
 
 | |
1M1Q
 
 | | P222 oxidized structure of the tetraheme cytochrome c from Shewanella oneidensis MR1 | | Descriptor: | HEME C, SULFATE ION, small tetraheme cytochrome c | | Authors: | Leys, D, Meyer, T.E, Tsapin, A.I, Nealson, K.H, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structures at atomic resolution reveal the novel concept of 'electron-harvesting' as a role for the small tetraheme cytochrome c
J.Biol.Chem., 277, 2002
|
|
1PJ6
 
 | | Crystal structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with folic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FOLIC ACID, N,N-dimethylglycine oxidase, ... | | Authors: | Leys, D, Basran, J, Scrutton, N.S. | | Deposit date: | 2003-06-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
Embo J., 22, 2003
|
|
1N4G
 
 | | Structure of CYP121, a Mycobacterial P450, in Complex with Iodopyrazole | | Descriptor: | 4-IODOPYRAZOLE, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D, Mowat, C.G, McLean, K.J, Richmond, A, Chapman, S.K, Walkinshaw, M.D, Munro, A.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-10-31 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic structure of Mycobacterium tuberculosis CYP121 to 1.06 A reveals novel features of cytochrome P450.
J.Biol.Chem., 278, 2003
|
|
3CXZ
 
 | |
3CY0
 
 | |
3CXY
 
 | |
3CY1
 
 | |
4L54
 
 | | Structure of cytochrome P450 OleT, ligand-free | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Terminal olefin-forming fatty acid decarboxylase | | Authors: | Leys, D. | | Deposit date: | 2013-06-10 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Biochemical Properties of the Alkene Producing Cytochrome P450 OleTJE (CYP152L1) from the Jeotgalicoccus sp. 8456 Bacterium.
J.Biol.Chem., 289, 2014
|
|
4L40
 
 | | Structure of the P450 OleT with a C20 fatty acid substrate bound | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Terminal olefin-forming fatty acid decarboxylase, icosanoic acid | | Authors: | Leys, D. | | Deposit date: | 2013-06-07 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Biochemical Properties of the Alkene Producing Cytochrome P450 OleTJE (CYP152L1) from the Jeotgalicoccus sp. 8456 Bacterium.
J.Biol.Chem., 289, 2014
|
|
3CXV
 
 | |
3CXX
 
 | |
3EKB
 
 | |
4KEW
 
 | | structure of the A82F BM3 heme domain in complex with omeprazole | | Descriptor: | 6-methoxy-2-{[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]sulfanyl}-1H-benzimidazole, Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Key Mutations Alter the Cytochrome P450 BM3 Conformational Landscape and Remove Inherent Substrate Bias.
J.Biol.Chem., 288, 2013
|
|
4KEY
 
 | | Structure of P450 BM3 A82F F87V in complex with omeprazole | | Descriptor: | 6-methoxy-2-{[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]sulfanyl}-1H-benzimidazole, Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Key Mutations Alter the Cytochrome P450 BM3 Conformational Landscape and Remove Inherent Substrate Bias.
J.Biol.Chem., 288, 2013
|
|
2FWT
 
 | | Crystal structure of DHC purified from Rhodobacter sphaeroides | | Descriptor: | DHC, diheme cytochrome c, HEME C | | Authors: | Leys, D. | | Deposit date: | 2006-02-03 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Studies on DHC, the Diheme Cytochrome c from Rhodobacter sphaeroides, and Its Interaction with SHP, the sphaeroides Heme Protein
Biochemistry, 45, 2006
|
|
4KF2
 
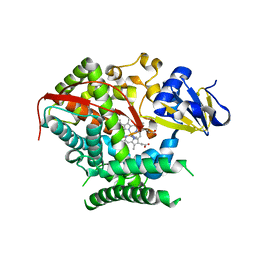 | | Structure of the P4509 BM3 A82F F87V heme domain | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Key Mutations Alter the Cytochrome P450 BM3 Conformational Landscape and Remove Inherent Substrate Bias.
J.Biol.Chem., 288, 2013
|
|
4KF0
 
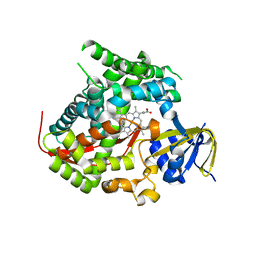 | | Structure of the A82F P450 BM3 heme domain | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Key Mutations Alter the Cytochrome P450 BM3 Conformational Landscape and Remove Inherent Substrate Bias.
J.Biol.Chem., 288, 2013
|
|
6TIO
 
 | |
6TIL
 
 | |
6TIC
 
 | |
6TIN
 
 | |
6TIH
 
 | |
6TIE
 
 | |
