3KK0
 
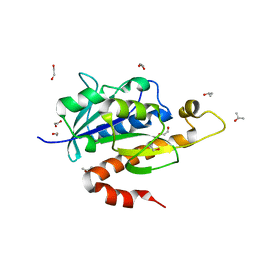 | | Crystal structure of partially folded intermediate state of peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | 分子名称: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kumar, A, Singh, N, Yadav, R, Sharma, S, Arora, A, Singh, T.P. | | 登録日 | 2009-11-04 | | 公開日 | 2010-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structures of Fully-Folded Native and Partially-Folded Intermediate States of Peptidyl-tRNA Hydrolase from Mycobacterium smegmatis
To be Published
|
|
7UCC
 
 | | Transcription factor FosB/JunD bZIP domain in the reduced form | | 分子名称: | CHLORIDE ION, ETHANOL, Protein fosB, ... | | 著者 | Kumar, A, Machius, M.C, Rudenko, G. | | 登録日 | 2022-03-16 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Chemically targeting the redox switch in AP1 transcription factor Delta FOSB.
Nucleic Acids Res., 50, 2022
|
|
7UCD
 
 | | Transcription factor FosB/JunD bZIP domain covalently modified with the cysteine-targeting alpha-haloketone compound Z2159931480 | | 分子名称: | 7-acetyl-4-methoxy-1-benzofuran-3(2H)-one, CHLORIDE ION, Protein fosB, ... | | 著者 | Kumar, A, Machius, M.C, Rudenko, G. | | 登録日 | 2022-03-16 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Chemically targeting the redox switch in AP1 transcription factor Delta FOSB.
Nucleic Acids Res., 50, 2022
|
|
4L69
 
 | |
7M6M
 
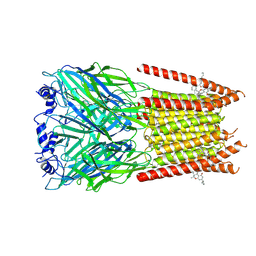 | | Full length alpha1 Glycine receptor in presence of 32uM Tetrahydrocannabinol | | 分子名称: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | 著者 | Kumar, A, Chakrapani, S. | | 登録日 | 2021-03-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6N
 
 | |
7M6Q
 
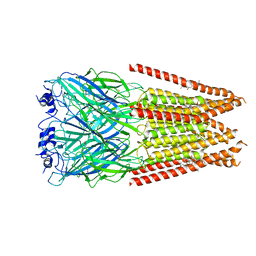 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 1 | | 分子名称: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | 著者 | Kumar, A, Chakrapani, S. | | 登録日 | 2021-03-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6R
 
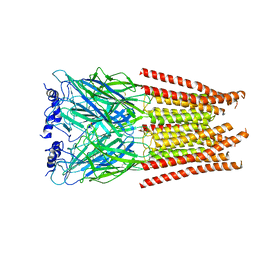 | |
7M6S
 
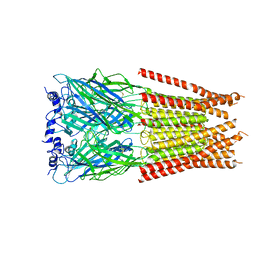 | |
7M6O
 
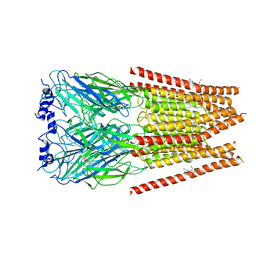 | | Full length alpha1 Glycine receptor in presence of 0.1mM Glycine and 32uM Tetrahydrocannabinol | | 分子名称: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | 著者 | Kumar, A, Chakrapani, S. | | 登録日 | 2021-03-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6P
 
 | |
8DK6
 
 | | Structure of hepatitis C virus envelope N-terminal truncated glycoprotein 2 (E2) (residues 456-713) from J6 genotype | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, 2A12 Fab light chain, ... | | 著者 | Kumar, A, Rohe, T, Elrod, E.J, Khan, A.G, Dearborn, A.D, Kissinger, R, Grakoui, A, Marcotrigiano, J. | | 登録日 | 2022-07-03 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Regions of hepatitis C virus E2 required for membrane association.
Nat Commun, 14, 2023
|
|
1HXI
 
 | | AN UNEXPECTED EXTENDED CONFORMATION FOR THE THIRD TPR MOTIF OF THE PEROXIN PEX5 FROM TRYPANOSOMA BRUCEI | | 分子名称: | MAGNESIUM ION, PEROXISOME TARGETING SIGNAL 1 RECEPTOR PEX5 | | 著者 | Kumar, A, Roach, C, Hirsh, I.S, Turley, S, deWalque, S, Michels, P.A.M, Hol, W.G.J. | | 登録日 | 2001-01-15 | | 公開日 | 2001-03-21 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | An unexpected extended conformation for the third TPR motif of the peroxin PEX5 from Trypanosoma brucei.
J.Mol.Biol., 307, 2001
|
|
3ND8
 
 | | Structural characterization for the nucleotide binding ability of subunit A of the A1AO ATP synthase | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ... | | 著者 | Kumar, A, Gruber, G. | | 登録日 | 2010-06-07 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The transition-like state and Pi entrance into the catalytic a subunit of the biological engine A-ATP synthase.
J.Mol.Biol., 408, 2011
|
|
6UD3
 
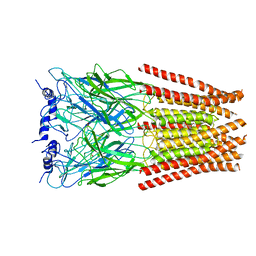 | | Full length Glycine receptor reconstituted in lipid nanodisc in Gly/PTX-bound open/blocked conformation | | 分子名称: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | 著者 | Kumar, A, Basak, S, Chakrapani, S. | | 登録日 | 2019-09-18 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
7MWX
 
 | | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2 with tamarin CD81 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy Chain, ... | | 著者 | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | 登録日 | 2021-05-17 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.32 Å) | | 主引用文献 | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
7MWS
 
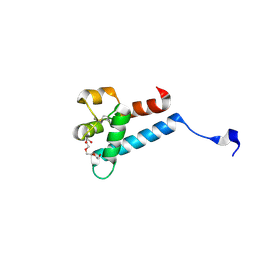 | | Crystal structure of tamarin CD81 large extracellular loop | | 分子名称: | CD81 protein, GLYCEROL, TETRAETHYLENE GLYCOL | | 著者 | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | 登録日 | 2021-05-17 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
7MWW
 
 | | Structure of hepatitis C virus envelope full-length glycoprotein 2 (eE2) from J6 genotype | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, ... | | 著者 | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | 登録日 | 2021-05-17 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
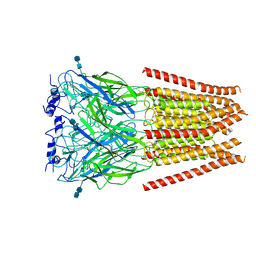
6UBT
 
 | |
7CXA
 
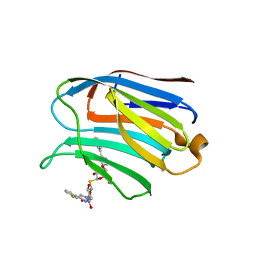 | | Structure of human Galectin-3 CRD in complex with TD-139 belonging to P31 space group. | | 分子名称: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | 著者 | Kumar, A. | | 登録日 | 2020-09-01 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7CXC
 
 | |
7CXB
 
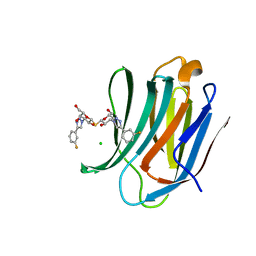 | | Structure of mouse Galectin-3 CRD in complex with TD-139 belonging to P6522 space group. | | 分子名称: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | 著者 | Kumar, A. | | 登録日 | 2020-09-01 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7CXD
 
 | |
6UBS
 
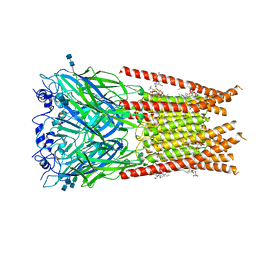 | | Full length Glycine receptor reconstituted in lipid nanodisc in Apo/Resting conformation | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1, ... | | 著者 | Kumar, A, Basak, S, Chakrapani, S. | | 登録日 | 2019-09-12 | | 公開日 | 2020-07-29 | | 最終更新日 | 2020-08-12 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
7VRR
 
 | |
