5SFH
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH N(C1CC1)(C)c2nn(c(n2)CCc4nn3c(cnc(c3n4)C)C)C, micromolar IC50=0.042255 | | Descriptor: | MAGNESIUM ION, N-cyclopropyl-5-{2-[(4S)-5,8-dimethyl[1,2,4]triazolo[1,5-a]pyrazin-2-yl]ethyl}-N,1-dimethyl-1H-1,2,4-triazol-3-amine, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SEF
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(nn(c(n1)CCc2nn3c(n2)c(c(nc3C)C)C)C)N4CCCC4, micromolar IC50=0.013626 | | Descriptor: | (4R)-5,7,8-trimethyl-2-{2-[1-methyl-3-(pyrrolidin-1-yl)-1H-1,2,4-triazol-5-yl]ethyl}[1,2,4]triazolo[1,5-c]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Lerner, C, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SEV
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(c(C)nc(c(n1)CCc2nc(cn2C)c3ccccc3)C)C, micromolar IC50=0.081773 | | Descriptor: | 2,3,5-trimethyl-6-{2-[(4S)-1-methyl-4-phenyl-4,5-dihydro-1H-imidazol-2-yl]ethyl}pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFI
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n1(nc(nc1CCc2nn3c(n2)c(ncc3C)C)N4[C@H](CCC4)C)C, micromolar IC50=0.028433 | | Descriptor: | (4S)-5,8-dimethyl-2-(2-{1-methyl-3-[(2S)-2-methylpyrrolidin-1-yl]-1H-1,2,4-triazol-5-yl}ethyl)[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFX
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2(c(c(C(=O)N1CCC1)cn2)C(Nc3ccc4c(c3)nc(s4)NC(C)=O)=O)C, micromolar IC50=0.012183 | | Descriptor: | MAGNESIUM ION, N-(2-acetamido-1,3-benzothiazol-5-yl)-4-(azetidine-1-carbonyl)-1-methyl-1H-pyrazole-5-carboxamide, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SF6
 
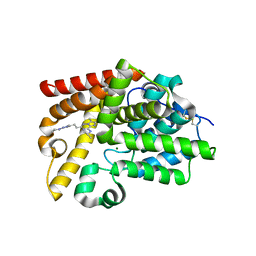 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH N1(CCCC1)c2nn(c(n2)CCc4nn3c(nc(c3C)C)c(n4)NC(C)C)C, micromolar IC50=0.002775 | | Descriptor: | (8S)-6,7-dimethyl-2-{2-[1-methyl-3-(pyrrolidin-1-yl)-1H-1,2,4-triazol-5-yl]ethyl}-N-(propan-2-yl)imidazo[2,1-f][1,2,4]triazin-4-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SE7
 
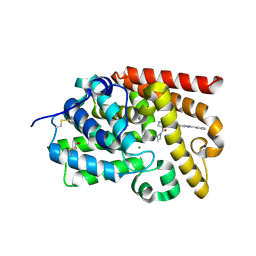 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2(c(c(C(=O)N1CCC1)cn2)C(Nc4cc3nc(nn3cc4)c5ccccc5)=O)C, micromolar IC50=0.00054035 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[(4S)-2-phenyl[1,2,4]triazolo[1,5-a]pyridin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFQ
 
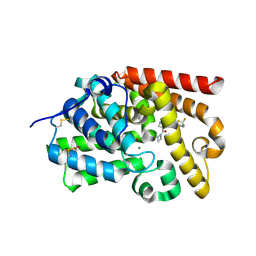 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2(c(c(C(=O)N1CCC1)cn2)C(Nc3nn(CC(F)F)cc3)=O)C, micromolar IC50=0.274828 | | Descriptor: | 4-(azetidine-1-carbonyl)-N-[1-(2,2-difluoroethyl)-1H-pyrazol-3-yl]-1-methyl-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Peters, J.U, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
1H8B
 
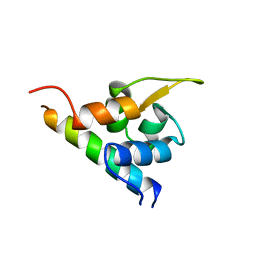 | | EF-hands 3,4 from alpha-actinin / Z-repeat 7 from titin | | Descriptor: | ALPHA-ACTININ 2, SKELETAL MUSCLE ISOFORM, TITIN | | Authors: | Atkinson, R.A, Joseph, C, Kelly, G, Muskett, F.W, Frenkiel, T.A, Nietlispach, D, Pastore, A. | | Deposit date: | 2001-02-01 | | Release date: | 2001-08-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ca2+-Independent Binding of an EF-Hand Domain to a Novel Motif in the Alpha-Actinin-Titin Complex
Nat.Struct.Biol., 8, 2001
|
|
4MJO
 
 | | Human liver fructose-1,6-bisphosphatase(d-fructose-1,6-bisphosphate, 1-phosphohydrolase) (e.c.3.1.3.11) complexed with the allosteric inhibitor 3 | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-({4-bromo-6-[(methylcarbamoyl)amino]pyridin-2-yl}carbamoyl)-5-(2-methoxyethyl)-4-methylthiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Tetaz, T, Benz, J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of protein-ligand binding constants of a cooperatively regulated tetrameric enzyme using electrospray mass spectrometry.
Acs Chem.Biol., 9, 2014
|
|
4AFS
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, FYNOMER, GLYCEROL, ... | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
4AG1
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, FYNOMER, SULFATE ION | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
4AFZ
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, D(-)-TARTARIC ACID, FYNOMER | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
1RGW
 
 | | Solution Structure of ZASP's PDZ domain | | Descriptor: | ZASP protein | | Authors: | Au, Y, Atkinson, R.A, Pallavicini, A, Joseph, C, Martin, S.R, Muskett, F.W, Guerrini, R, Faulkner, G, Pastore, A. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of ZASP PDZ Domain; Implications for Sarcomere Ultrastructure and Enigma Family Redundancy.
Structure, 12, 2004
|
|
4AFQ
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHYMASE, CITRATE ANION, ... | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
1VIH
 
 | | NMR STUDY OF VIGILIN, REPEAT 6, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
1VIG
 
 | | NMR STUDY OF VIGILIN, REPEAT 6, 40 STRUCTURES | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
4AFU
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, FYNOMER | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
4AG2
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHYMASE, ... | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
2Y5K
 
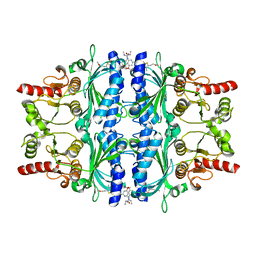 | | Orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | Descriptor: | 1-[5-(2-METHOXYETHYL)-4-METHYL-THIOPHEN-2-YL]SULFONYL-3-[4-METHOXY-6-(METHYLCARBAMOYLAMINO)PYRIDIN-2-YL]UREA, FRUCTOSE-1,6-BISPHOSPHATASE 1 | | Authors: | Ruf, A, Hebeisen, P, Haap, W, Kuhn, B, Mohr, P, Wessel, H.P, Zutter, U, Kirchner, S, Benz, J, Joseph, C, Alvarez-Sanchez, R, Gubler, M, Schott, B, Benardeau, A, Tozzo, E, Kitas, E. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y5L
 
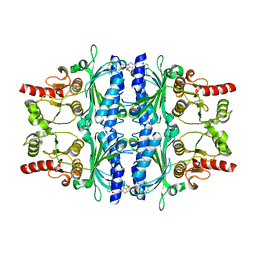 | | orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-{[(2Z)-5-bromo-1,3-thiazol-2(3H)-ylidene]carbamoyl}-3-chlorobenzenesulfonamide | | Authors: | ruf, a, hebeisen, p, haap, w, kuhn, b, mohr, p, wessel, h.p, zutter, u, kirchner, s, benz, j, joseph, c, alvarez-sanchez, r, gubler, m, schott, b, benardeau, a, tozzo, e, kitas, e. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2WBD
 
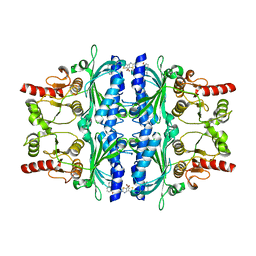 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE-1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH AN AMP SITE INHIBITOR | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-3-ethylbenzenesulfonamide | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T, Kitas, E, Mohr, P, Kuhn, B, Wessel, H.P, Hebeisen, P, Haap, W, Huber, W, Alvarez Sanchez, R, Paehler, A, Bernadeau, A, Gubler, M, Schott, B, Tozzo, E. | | Deposit date: | 2009-02-26 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sulfonylureido Thiazoles as Fructose-1,6-Bisphosphatase Inhibitors for the Treatment of Type-2 Diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2VT5
 
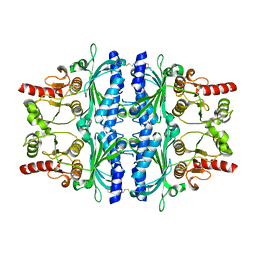 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE -1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH A DUAL BINDING AMP SITE INHIBITOR | | Descriptor: | 4-AMINO-N-[(2-SULFANYLETHYL)CARBAMOYL]BENZENESULFONAMIDE, FRUCTOSE-1,6-BISPHOSPHATASE 1 | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric Fbpase Inhibitors Gain 10(5) Times in Potency When Simultaneously Binding Two Neighboring AMP Sites.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2WBB
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE-1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH AN AMP SITE INHIBITOR | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-{[(2Z)-5-BROMO-1,3-THIAZOL-2(3H)-YLIDENE]CARBAMOYL}-4-METHYLBENZENESULFONAMIDE | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T, Kitas, E, Mohr, P, Kuhn, B, Wessel, H.P, Hebeisen, P, Haap, W, Huber, W, Alvarez Sanchez, R, Paehler, A, Bernadeau, A, Gubler, M, Schott, B, Tozzo, E. | | Deposit date: | 2009-02-26 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Sulfonylureido Thiazoles as Fructose-1,6-Bisphosphatase Inhibitors for the Treatment of Type-2 Diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3S3X
 
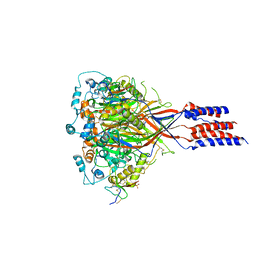 | | Structure of chicken acid-sensing ion channel 1 AT 3.0 A resolution in complex with psalmotoxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive cation channel 2, neuronal, ... | | Authors: | Dawson, R.J.P, Benz, J, Stohler, P, Tetaz, T, Joseph, C, Huber, S, Schmid, G, Huegin, D, Pflimlin, P, Trube, G, Rudolph, M.G, Hennig, M, Ruf, A. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of the Acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1.
Nat Commun, 3, 2012
|
|
