2MYS
 
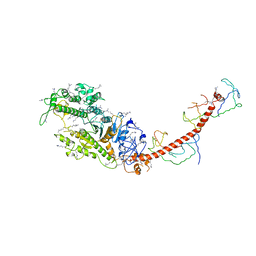 | |
6V33
 
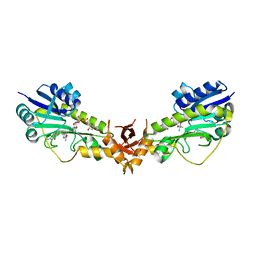 | | X-ray structure of a sugar N-formyltransferase from Pseudomonas congelans | | Descriptor: | 1,2-ETHANEDIOL, FOLIC ACID, dTDP-4-amino-4,6-dideoxyglucose, ... | | Authors: | Girardi, N.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-11-25 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Misannotations of the genes encoding sugar N-formyltransferases.
Protein Sci., 29, 2020
|
|
6V2T
 
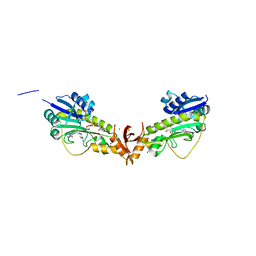 | | X-ray structure of a sugar N-formyltransferase from Shewanella sp FDAARGOS_354 | | Descriptor: | 1,2-ETHANEDIOL, FOLIC ACID, PHOSPHATE ION, ... | | Authors: | Girardi, N.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-11-25 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Misannotations of the genes encoding sugar N-formyltransferases.
Protein Sci., 29, 2020
|
|
6VLO
 
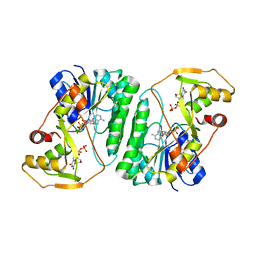 | | X-ray Structure of the R141 Sugar 4,6-dehydratase from Acanthamoeba polyphaga Minivirus | | Descriptor: | NICKEL (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative dTDP-D-glucose 4,6-dehydratase, ... | | Authors: | Thoden, J.B, Ferek, J.D, Holden, H.M. | | Deposit date: | 2020-01-24 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical analysis of a sugar 4,6-dehydratase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 29, 2020
|
|
6VO8
 
 | | X-ray structure of the Cj1427 in the presence of NADH and GDP-D-glycero-D-mannoheptose, an essential NAD-dependent dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative sugar-nucleotide epimerase/dehydratease, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S},5~{S},6~{S})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Spencer, K.D, Anderson, T.K, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
6VO6
 
 | | Crystal Structure of Cj1427, an Essential NAD-dependent Dehydrogenase from Campylobacter jejuni, in the Presence of NADH and GDP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Anderson, T.K, Spencer, K.D, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
6B5E
 
 | | Mycobacterium tuberculosis RmlA in complex with dTDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, CHLORIDE ION, ... | | Authors: | Brown, H.A, Holden, H.M. | | Deposit date: | 2017-09-29 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of glucose-1-phosphate thymidylyltransferase from Mycobacterium tuberculosis reveals the location of an essential magnesium ion in the RmlA-type enzymes.
Protein Sci., 27, 2018
|
|
6CBO
 
 | | X-ray structure of GenB1 from micromonospora echinospora in complex with neamine and PLP (as the external aldimine) | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranoside, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Dow, G.T, Thoden, J.B, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
6CBK
 
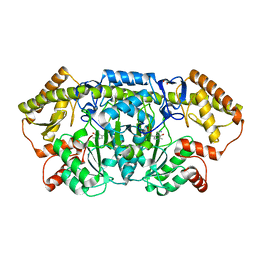 | | X-ray structure of NeoB from Streptomyces fradiae in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Neamine transaminase NeoN, ... | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
6CBL
 
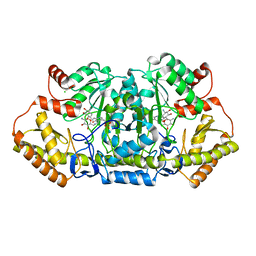 | | x-ray structure of NeoB from Streptomyces fradiae in complex with neamine as an external aldimine | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranoside, CHLORIDE ION, Neamine transaminase NeoN | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
6CBM
 
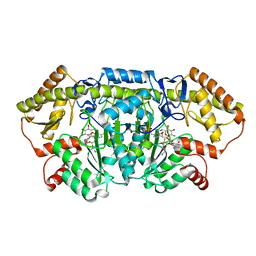 | | x-ray structure of NeoB from streptomyces fradiae in complex with PLP and neomycin (as the external aldimine) at pH 9 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-[(3-O-{2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranosyl}-beta-D-ribofuranosyl)oxy]-3-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
6CBN
 
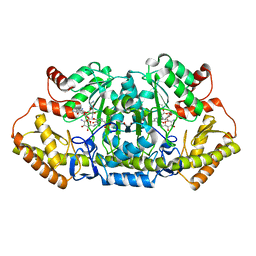 | | x-ray structure of NeoB from streptomyces fradiae in complex with PLP and neomycin (as the external aldimine) at pH 7.5 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-[(3-O-{2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranosyl}-beta-D-ribofuranosyl)oxy]-3-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 1,2-ETHANEDIOL, Neamine transaminase NeoN | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
6BWL
 
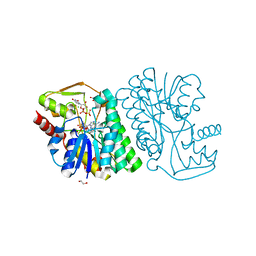 | | X-ray structure of Pal from Bacillus thuringiensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Delvaux, N.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular architectures of Pen and Pal: Key enzymes required for CMP-pseudaminic acid biosynthesis in Bacillus thuringiensis.
Protein Sci., 27, 2018
|
|
6BWC
 
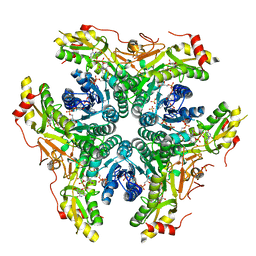 | | X-ray structure of Pen from Bacillus thuringiensis | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Polysaccharide biosynthesis protein CapD, SULFATE ION, ... | | Authors: | Delvaux, N.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-12-14 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular architectures of Pen and Pal: Key enzymes required for CMP-pseudaminic acid biosynthesis in Bacillus thuringiensis.
Protein Sci., 27, 2018
|
|
3NDI
 
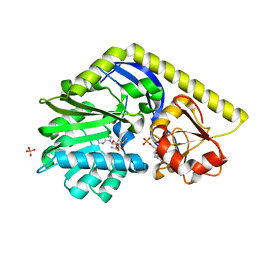 | | X-ray Structure of a C-3'-Methyltransferase in Complex with S-adenosylmethionine and dTMP | | Descriptor: | Methyltransferase, PHOSPHATE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Bruender, N.A, Thoden, J.B, Kaur, M, Avey, M.K, Holden, H.M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Architecture of a C-3'-Methyltransferase Involved in the Biosynthesis of d-Tetronitrose.
Biochemistry, 49, 2010
|
|
5BJX
 
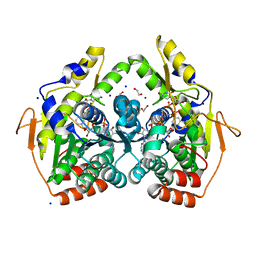 | | X-ray structure of the PglF 4,6-dehydratase from campylobacter jejuni, variant T395V, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5BJY
 
 | | x-ray structure of the PglF 4,5-dehydratase from campylobacter jejuni, variant M405Y, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5BSZ
 
 | | X-ray structure of the sugar N-methyltransferase KedS8 from Streptoalloteichus sp ATCC 53650 | | Descriptor: | 1,2-ETHANEDIOL, N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Thoden, J.B, Delvaux, N.A, Holden, H.M. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular architecture of KedS8, a sugar N-methyltransferase from Streptoalloteichus sp. ATCC 53650.
Protein Sci., 24, 2015
|
|
7L82
 
 | | x-ray structure of the psychrobacter cryohalolentis Pcryo_0637 N-acetyltransferase in the presene of its reaction tetrahedral intermediate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Putative acetyl transferase protein, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
7L7Y
 
 | | x-ray structure of the N-acetyltransferase Pcryo_0637 from psychrobacter cryohalolentis in the presence of UDP and acetyl-conezyme A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
7L7Z
 
 | | x-ray structure of the N-acetyltransferase Pcryo_0637 from psychrobacter cryohalolentis in the presence of coenzyme A and UDP-di-N-acetyl-bacillosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
7L81
 
 | | x-ray structure of the psychrobacter cryohalolentis N-acetyltransferase Pcryo_0637 in the presence of coenzyme A and | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
7L7X
 
 | | X-ray structure of the Pcryo_0638 aminotransferase from Psychrobacter cryohalolentis | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DegT/DnrJ/EryC1/StrS aminotransferase, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
3NDJ
 
 | | X-ray Structure of a C-3'-Methyltransferase in Complex with S-Adenosyl-L-Homocysteine and Sugar Product | | Descriptor: | (2R,4S,6R)-4-amino-4,6-dimethyl-5-oxotetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), Methyltransferase, PHOSPHATE ION, ... | | Authors: | Bruender, N.A, Thoden, J.B, Kaur, M, Avey, M.K, Holden, H.M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Architecture of a C-3'-Methyltransferase Involved in the Biosynthesis of d-Tetronitrose.
Biochemistry, 49, 2010
|
|
3FRK
 
 | | X-ray structure of QdtB from T. thermosaccharolyticum in complex with a PLP:TDP-3-aminoquinovose aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, QdtB | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of QdtB, an aminotransferase required for the biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-glucose.
Biochemistry, 48, 2009
|
|
