9AT8
 
 | | Fab 77-stabilized MeV F ectodomain fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, mAB 77 heavy chain, ... | | Authors: | Zyla, D, Saphire, E.O. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A neutralizing antibody prevents postfusion transition of measles virus fusion protein.
Science, 384, 2024
|
|
5LP9
 
 | | FimA wt from S. flexneri | | Descriptor: | Major type 1 subunit fimbrin (Pilin) | | Authors: | Zyla, D, Capitani, G, Prota, A, Glockshuber, R. | | Deposit date: | 2016-08-12 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.88626635 Å) | | Cite: | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 2019
|
|
6ERJ
 
 | | Self-complemented FimA subunit from Salmonella enterica | | Descriptor: | ACETIC ACID, Type-1 fimbrial protein, a chain | | Authors: | Zyla, D.S, Prota, A, Capitani, G, Glockshuber, R. | | Deposit date: | 2017-10-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 294, 2019
|
|
8PTU
 
 | | 2.5 A cryo-EM structure of the in vitro FimD-catalyzed assembly of type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Hospenthal, M, Glockshuber, R, Waksman, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
6Y7S
 
 | | 2.85 A cryo-EM structure of the in vivo assembled type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Hospenthal, M, Waksman, G, Glockshuber, R. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-31 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
6R74
 
 | | N-terminally reversed variant of FimA E. coli | | Descriptor: | SULFATE ION, Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Echeverria, B, Glockshuber, R. | | Deposit date: | 2019-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Donor strand sequence, rather than donor strand orientation, determines the stability and non-equilibrium folding of the type 1 pilus subunit FimA.
J.Biol.Chem., 295, 2020
|
|
6R7E
 
 | | N-terminally reversed variant of FimA E. coli with alanine insertion at position 20 | | Descriptor: | FimA, SULFATE ION | | Authors: | Zyla, D, Echeverria, B, Glockshuber, R. | | Deposit date: | 2019-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Donor strand sequence, rather than donor strand orientation, determines the stability and non-equilibrium folding of the type 1 pilus subunit FimA.
J.Biol.Chem., 295, 2020
|
|
6S09
 
 | | C-terminally extended and N-terminally truncated variant of FimA E. coli at 1.5 Angstrom resolution | | Descriptor: | ACETIC ACID, FimA, SODIUM ION, ... | | Authors: | Zyla, D, Echeverria, B, Glockshuber, R. | | Deposit date: | 2019-06-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Donor strand sequence, rather than donor strand orientation, determines the stability and non-equilibrium folding of the type 1 pilus subunit FimA.
J.Biol.Chem., 295, 2020
|
|
8UTF
 
 | |
8UUP
 
 | | Structure of the Measles virus Fusion protein in the pre-fusion conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Zyla, D, Saphire, E.O. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | A neutralizing antibody prevents postfusion transition of measles virus fusion protein.
Science, 384, 2024
|
|
8UT2
 
 | | Pre-fusion Measles virus fusion protein complexed with Fab 77 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zyla, D, Saphire, E.O. | | Deposit date: | 2023-10-30 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | A neutralizing antibody prevents postfusion transition of measles virus fusion protein.
Science, 384, 2024
|
|
8UUQ
 
 | |
5NKT
 
 | | FimA wt from E. coli | | Descriptor: | SULFATE ION, Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Capitani, G, Prota, A, Glockshuber, R. | | Deposit date: | 2017-04-03 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 2019
|
|
6ZS5
 
 | | 3.5 A cryo-EM structure of human uromodulin filament core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
6ZYA
 
 | | Extended human uromodulin filament core at 3.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
8PSV
 
 | | 2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Hospenthal, M, Zyla, D, Glockshuber, R, Waksman, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
6X84
 
 | |
8F0G
 
 | | Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibody Fab 1C3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 1C3 Fab Heavy Chain, ... | | Authors: | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
8F0H
 
 | | Structure of SARS-CoV-2 spike with antibody Fabs 2A10 and 1H2 (Local refinement of the RBD and Fabs 1H2 and 2A10) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 1H2 heavy chain, Antibody Fab 1H2 light chain, ... | | Authors: | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
6SYM
 
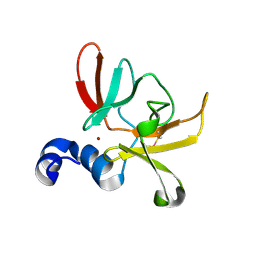 | |
6YEV
 
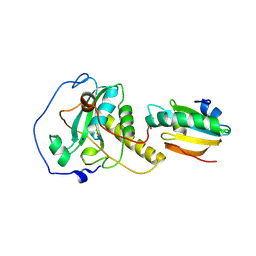 | |
7U9G
 
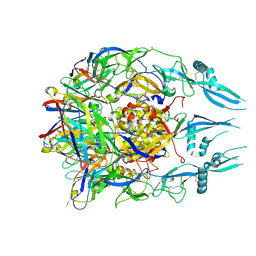 | | Rabies virus glycoprotein pre-fusion trimer in complex with neutralizing antibody RVA122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, RVA122 Fab Heavy Chain, ... | | Authors: | Callaway, H.M, Zyla, D, Larrous, F, Dias de Melo, G, Hastie, K.M, Avalos, R.D, Agarwal, A, Bouhry, H, Corti, D, Saphire, E.O. | | Deposit date: | 2022-03-10 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structure of the rabies virus glycoprotein trimer bound to a prefusion-specific neutralizing antibody.
Sci Adv, 8, 2022
|
|
8DMI
 
 | | Lymphocytic choriomeningitis virus glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein G1, ... | | Authors: | Moon-Walker, A, Hastie, K.M, Zyla, D.S, Saphire, E.O. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for antibody-mediated neutralization of lymphocytic choriomeningitis virus.
Cell Chem Biol, 30, 2023
|
|
8DMH
 
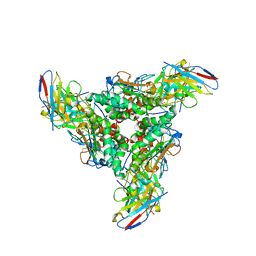 | | Lymphocytic choriomeningitis virus glycoprotein in complex with neutralizing antibody M28 | | Descriptor: | 18.5C-M28 Fab Heavy Chain, 18.5C-M28 Fab Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Moon-Walker, A, Hastie, K.M, Zyla, D.S, Saphire, E.O. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for antibody-mediated neutralization of lymphocytic choriomeningitis virus.
Cell Chem Biol, 30, 2023
|
|
8USN
 
 | | Intracellular cryo-tomography structure of EBOV nucleocapsid at 8.9 Angstrom | | Descriptor: | Membrane-associated protein VP24, Nucleoprotein, Polymerase cofactor VP35, ... | | Authors: | Watanabe, R, Zyla, D, Saphire, E.O. | | Deposit date: | 2023-10-27 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Intracellular Ebola virus nucleocapsid assembly revealed by in situ cryo-electron tomography.
Cell, 187, 2024
|
|
