4E6U
 
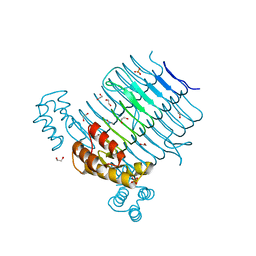 | | Structure of LpxA from Acinetobacter baumannii at 1.4A resolution (P63 form) | | Descriptor: | 1,2-ETHANEDIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, SULFATE ION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-16 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4E6T
 
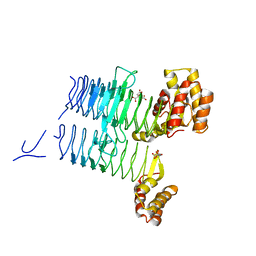 | | Structure of LpxA from Acinetobacter baumannii at 1.8A resolution (P212121 form) | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CITRATE ANION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-15 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4E75
 
 | | Structure of LpxD from Acinetobacter baumannii at 2.85A resolution (P21 form) | | Descriptor: | UDP-3-O-acylglucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-16 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure determination of LpxD from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5U35
 
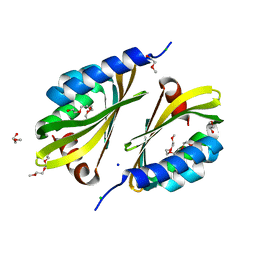 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-METHOXYETHANOL, CHLORIDE ION, ... | | Authors: | Oberdorfer, G, Marcos, E, Basanta, B, Chidyausiku, T.M, Sankaran, B, Zwart, P.H, Baker, D. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
3QI0
 
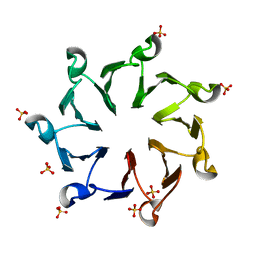 | | Structural, thermodynamic and kinetic analysis of the picomolar binding affinity interaction of the beta-lactamase inhibitor protein-II (BLIP-II) with class A beta-lactamases | | Descriptor: | Beta-lactamase inhibitory protein II, SULFATE ION | | Authors: | Brown, N.G, Chow, D.C, Sankaran, B, Zwart, P, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2011-01-26 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the binding forces driving the tight interactions between beta-lactamase inhibitory protein-II (BLIP-II) and class A beta-lactamases.
J.Biol.Chem., 286, 2011
|
|
5TS4
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Chidyausiku, T.M, Marcos, E, Pereira, J.H, Sankaran, B, Zwart, P.H, Baker, D. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
3OGJ
 
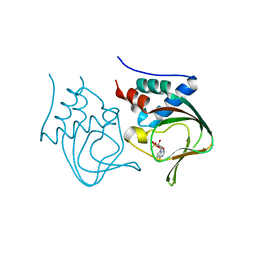 | | Crystal structure of partial apo (92-227) of cGMP-dependent protein kinase | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PHOSPHATE ION, PRKG1 protein | | Authors: | Kim, J.J, Huang, G, Kwon, T.K, Zwart, P, Headd, J, Kim, C. | | Deposit date: | 2010-08-16 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Co-Crystal Structures of PKG Ibeta (92-227) with cGMP and cAMP Reveal the Molecular Details of Cyclic-Nucleotide Binding
Plos One, 6, 2011
|
|
3OCP
 
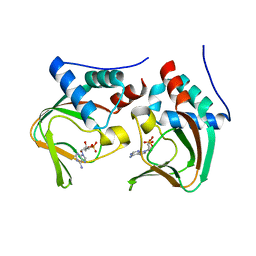 | | Crystal structure of cAMP bound cGMP-dependent protein kinase(92-227) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PRKG1 protein | | Authors: | Kim, J.J, Huang, G, Kwon, T.K, Zwart, P, Headd, J, Kim, C. | | Deposit date: | 2010-08-10 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Co-Crystal Structures of PKG Ibeta (92-227) with cGMP and cAMP Reveal the Molecular Details of Cyclic-Nucleotide Binding
Plos One, 6, 2011
|
|
3OD0
 
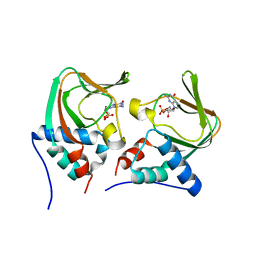 | | Crystal structure of cGMP bound cGMP-dependent protein kinase(92-227) | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, PRKG1 protein | | Authors: | Kim, J.J, Huang, G, Kwon, T.K, Zwart, P, Headd, J, Kim, C. | | Deposit date: | 2010-08-10 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Co-Crystal Structures of PKG Ibeta (92-227) with cGMP and cAMP Reveal the Molecular Details of Cyclic-Nucleotide Binding
Plos One, 6, 2011
|
|
3MG2
 
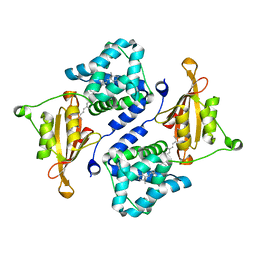 | | Crystal structure of the orange carotenoid protein Y44S mutant from cyanobacteria synechocystis sp. PCC 6803 | | Descriptor: | Orange carotenoid protein, beta,beta-caroten-4-one | | Authors: | Wilson, A, Kinney, J, Zwart, P.H, Punginelli, C, D'Haen, S, Perreau, F, Klein, M.G, Kirilovsky, D, Kerfeld, C.A. | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria.
J.Biol.Chem., 285, 2010
|
|
3MG1
 
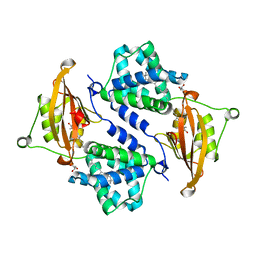 | | Crystal structure of the orange carotenoid protein from cyanobacteria Synechocystis sp. PCC 6803 | | Descriptor: | GLYCEROL, Orange carotenoid protein, beta,beta-caroten-4-one | | Authors: | Wilson, A, Kinney, J, Zwart, P.H, Punginelli, C, D'Haen, S, Perreau, F, Klein, M.G, Kirilovsky, D, Kerfeld, C.A. | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria.
J.Biol.Chem., 285, 2010
|
|
3MG3
 
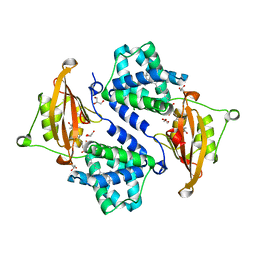 | | Crystal structure of the orange carotenoid protein R155L mutant from cyanobacteria synechocystis sp. PCC 6803 | | Descriptor: | GLYCEROL, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | Authors: | Wilson, A, Kinney, J, Zwart, P.H, Punginelli, C, D'Haen, S, Perreau, F, Klein, M.G, Kirilovsky, D, Kerfeld, C.A. | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria.
J.Biol.Chem., 285, 2010
|
|
3Q6L
 
 | | Crystal Structure of Human Adipocyte Fatty Acid Binding Protein (FABP4) at 1.4 Ang. Resolution | | Descriptor: | Fatty acid-binding protein, adipocyte | | Authors: | Prasad, G.S, Carney, D, Small, V, Sankaran, B, Zwart, P.H. | | Deposit date: | 2011-01-02 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Human Adipocyte Fatty Acid Binding Protein (FABP4) at 1.4 Ang. Resolution
To be Published
|
|
3PMO
 
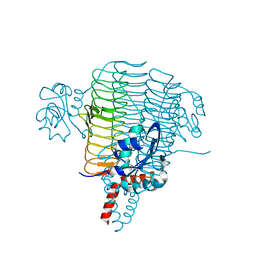 | | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2010-11-17 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5K7V
 
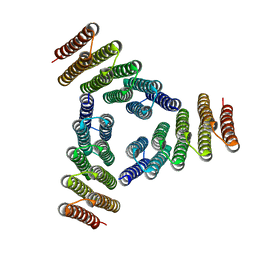 | | Computational Design of Self-Assembling Cyclic Protein Homooligomers | | Descriptor: | Designed protein HR00C3 | | Authors: | Sankaran, B, Zwart, P.H, Fallas, J.A, Pereira, J.H, Ueda, G, Baker, D. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.165 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5KBA
 
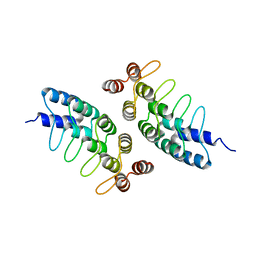 | | Computational Design of Self-Assembling Cyclic Protein Homooligomers | | Descriptor: | Designed protein ANK1C2 | | Authors: | Sankaran, B, Zwart, P.H, Fallas, J.A, Pereira, J.H, Ueda, G, Baker, D. | | Deposit date: | 2016-06-02 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5J0I
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | Designed protein 2L6HC3_12 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5JQZ
 
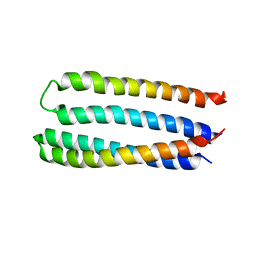 | | Designed two-ring homotetramer at 3.8A resolution | | Descriptor: | De novo designed homotetramer | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Oberdorfer, G, Boyken, S.E, Chen, Z. | | Deposit date: | 2016-05-05 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.83 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5JE7
 
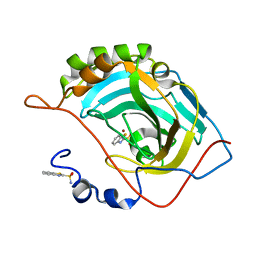 | | Human carbonic anhydrase II (F131Y) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5JG5
 
 | | Human carbonic anhydrase II (V121T/F131Y) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5JGT
 
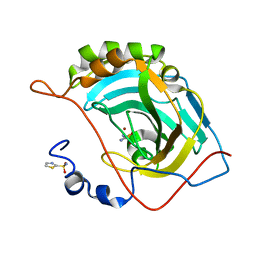 | | Human carbonic anhydrase II (F131Y/L198A) complexed with 1,3-thiazole-2-sulfonamide | | Descriptor: | 1,3-thiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5IZS
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | Designed protein 5L6HC3_1 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Oberdorfer, G. | | Deposit date: | 2016-03-25 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J0K
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | designed protein 2L4HC2_23 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
