6JUZ
 
 | | Crystal Structure of N-terminal domain of ArgZ(N71S) covalently bond to a reaction intermediate | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhuang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
4HU0
 
 | |
4HTY
 
 | |
5JK5
 
 | |
5JK6
 
 | |
5JK8
 
 | |
4HAC
 
 | |
6JV0
 
 | | Crystal Structure of N-terminal domain of ArgZ, bound to Product, an arginine dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | 1,2-ETHANEDIOL, L-ornithine, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
6JV1
 
 | | Crystal Structure of N-terminal domain of ArgZ, C264S mutant, bound to Substrate, an arginine dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | ARGININE, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
6JUY
 
 | | Crystal Structure of ArgZ, apo structure, an Arginine Dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
3UK7
 
 | |
3ORF
 
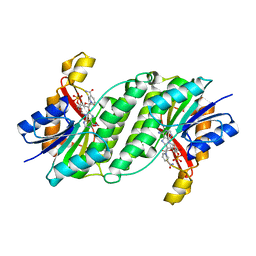 | | Crystal Structure of Dihydropteridine Reductase from Dictyostelium discoideum | | Descriptor: | Dihydropteridine reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chen, C, Zhuang, N.N, Seo, K.H, Park, Y.S, Lee, K.H. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural insights into the dual substrate specificities of mammalian and Dictyostelium dihydropteridine reductases toward two stereoisomers of quinonoid dihydrobiopterin
Febs Lett., 585, 2011
|
|
4DMM
 
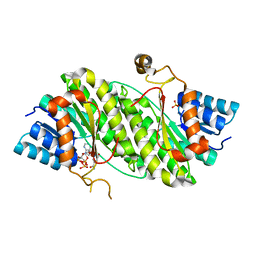 | |
4DML
 
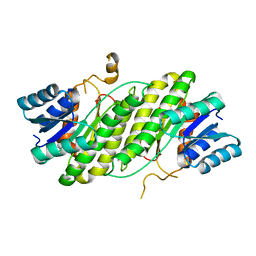 | |
8T7C
 
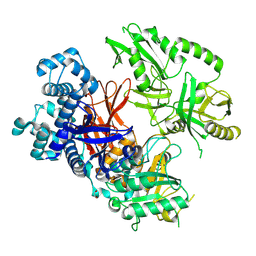 | | Crystal structure of human phospholipase C gamma 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2, CALCIUM ION | | Authors: | Chen, Y, Choi, H, Zhuang, N, Hu, L, Qian, D, Wang, J. | | Deposit date: | 2023-06-20 | | Release date: | 2024-06-26 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal and cryo-EM structures of PLC gamma 2 reveal dynamic interdomain recognitions in autoinhibition.
Sci Adv, 10, 2024
|
|
8UPS
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 5 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, PHOSPHATE ION | | Authors: | Wu, Y, Qiang, D, Zhuang, N, Krishnamurthy, H, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPW
 
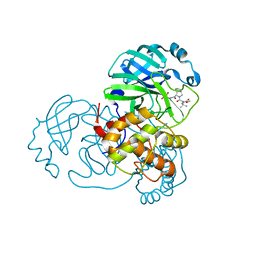 | | Structure of SARS-Cov2 3CLPro in complex with Compound 34 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6S)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPV
 
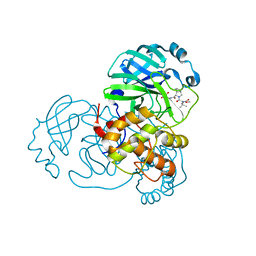 | | Structure of SARS-Cov2 3CLPro in complex with Compound 33 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6R)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UTE
 
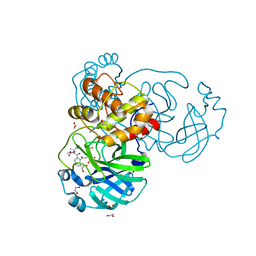 | | Structure of SARS-Cov2 3CLPro in complex with Compound 27 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S)-6,6-difluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-31 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
3QNA
 
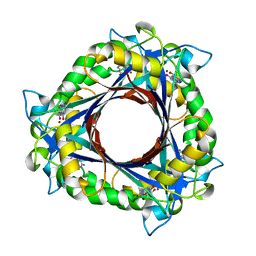 | | Crystal structure of a 6-pyruvoyltetrahydropterin synthase homologue from Esherichia coli complexed sepiapterin | | Descriptor: | 6-carboxy-5,6,7,8-tetrahydropterin synthase, BIOPTERIN, ZINC ION | | Authors: | Seo, K.H, Zhuang, N.N, Lee, K.H. | | Deposit date: | 2011-02-08 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of a novel activity of bacterial 6-pyruvoyltetrahydropterin synthase homologues distinct from mammalian 6-pyruvoyltetrahydropterin synthase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3QN9
 
 | |
3QN0
 
 | | Structure of 6-pyruvoyltetrahydropterin synthase | | Descriptor: | 6-carboxy-5,6,7,8-tetrahydropterin synthase, ZINC ION | | Authors: | Seo, K.H, Zhuang, N.N, Lee, K.H. | | Deposit date: | 2011-02-07 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis of a novel activity of bacterial 6-pyruvoyltetrahydropterin synthase homologues distinct from mammalian 6-pyruvoyltetrahydropterin synthase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3TWO
 
 | | The crystal structure of CAD from Helicobacter pylori complexed with NADP(H) | | Descriptor: | Mannitol dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Seo, K.H, Zhuang, N.N, Cong, C, Lee, K.H. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Unusual NADPH conformation in the crystal structure of a cinnamyl alcohol dehydrogenase from Helicobacter pylori in complex with NADP(H) and substrate docking analysis
Febs Lett., 586, 2012
|
|
