5DDZ
 
 | | Crystal structure of the RTA-c10-P2 complex | | Descriptor: | 60S acidic ribosomal protein P2, Ricin | | Authors: | Zhu, Y, Fan, X, Wang, C, Niu, L, Li, X, Teng, M. | | Deposit date: | 2015-08-25 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the interaction of the ribosomal P stalk protein P2 with a type II ribosome-inactivating protein ricin
Sci Rep, 6, 2016
|
|
6Y9N
 
 | | Crystal structure of Whirlin PDZ3_C-ter in complex with Myosin 15a C-terminal PDZ binding motif peptide | | Descriptor: | Unconventional myosin-XV, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
6Y9O
 
 | | Crystal structure of Whirlin PDZ3_C-ter in complex with CASK internal PDZ binding motif peptide | | Descriptor: | Peripheral plasma membrane protein CASK, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
6Y9Q
 
 | | Crystal structure of Whirlin PDZ3_C-ter in complex with Taperin internal PDZ binding motif peptide | | Descriptor: | Taperin, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.315 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
1F2S
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN BOVINE BETA-TRYPSIN AND MCTI-A, A TRYPSIN INHIBITOR OF SQUASH FAMILY AT 1.8 A RESOLUTION | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR A | | Authors: | Zhu, Y, Huang, Q, Qian, M, Jia, Y, Tang, Y. | | Deposit date: | 2000-05-29 | | Release date: | 2000-06-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the complex formed between bovine beta-trypsin and MCTI-A, a trypsin inhibitor of squash family, at 1.8-A resolution.
J.Protein Chem., 18, 1999
|
|
6Y9P
 
 | | Crystal structure of Whirlin PDZ3_C-ter in complex with Harmonin a1 C-terminal PDZ binding motif peptide | | Descriptor: | Harmonin a1, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.169 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
6Y38
 
 | | Crystal structure of Whirlin PDZ3 in complex with Myosin 15a C-terminal PDZ binding motif peptide | | Descriptor: | Chains: C,D, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-02-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
2WZS
 
 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 in complex with Mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhu, Y, Suits, M.D.L, Thompson, A, Chavan, S, Dinev, Z, Dumon, C, Smith, N, Moremen, K.W, Xiang, Y, Siriwardena, A, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-12-02 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
1XDP
 
 | | Crystal Structure of the E.coli Polyphosphate Kinase in complex with AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyphosphate kinase | | Authors: | Zhu, Y, Huang, W, Lee, S.S, Xu, W. | | Deposit date: | 2004-09-07 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a polyphosphate kinase and its implications for polyphosphate synthesis
Embo Rep., 6, 2005
|
|
1XDO
 
 | |
3CVR
 
 | | Crystal structure of the full length IpaH3 | | Descriptor: | Invasion plasmid antigen | | Authors: | Zhu, Y, Shao, F. | | Deposit date: | 2008-04-19 | | Release date: | 2008-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Shigella effector reveals a new class of ubiquitin ligases
Nat.Struct.Mol.Biol., 15, 2008
|
|
7QXX
 
 | | Proteasome-ZFAND5 Complex Z+E state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QXN
 
 | | Proteasome-ZFAND5 Complex Z+A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QY7
 
 | | Proteasome-ZFAND5 Complex Z-A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QYB
 
 | | Proteasome-ZFAND5 Complex Z-C state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QXP
 
 | | Proteasome-ZFAND5 Complex Z+B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QXU
 
 | | Proteasome-ZFAND5 Complex Z+C state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QXW
 
 | | Proteasome-ZFAND5 Complex Z+D state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QYA
 
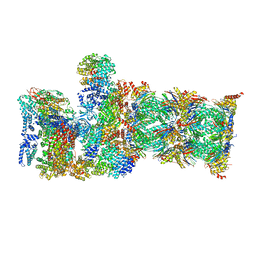 | | Proteasome-ZFAND5 Complex Z-B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
3L0I
 
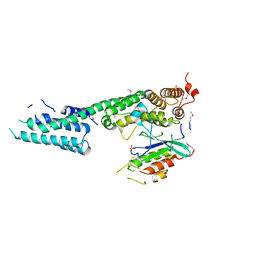 | | Complex structure of SidM/DrrA with the wild type Rab1 | | Descriptor: | CHLORIDE ION, DrrA, Ras-related protein Rab-1A, ... | | Authors: | Zhu, Y, Shao, F. | | Deposit date: | 2009-12-10 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural mechanism of host Rab1 activation by the bifunctional Legionella type IV effector SidM/DrrA
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3L0M
 
 | |
1MUJ
 
 | |
7QCT
 
 | | PDZ2 of LNX2 with SARS-CoV-2_E PBM complex | | Descriptor: | Envelope small membrane protein, Ligand of Numb protein X 2 | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
7QCR
 
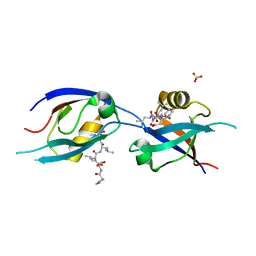 | | MLLT4/Afadin PDZ domain in complex with the C-terminal peptide from protein E of SARS-CoV-2 | | Descriptor: | Afadin, Envelope small membrane protein, SULFATE ION | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
7QCS
 
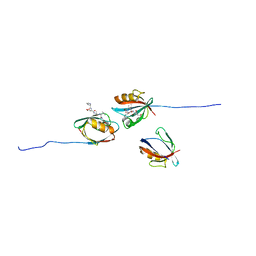 | | PALS1/MPP5 PDZ domain in complex with SARS-CoV-2_E PBM peptide | | Descriptor: | Envelope small membrane protein, Protein PALS1 | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
