8TKN
 
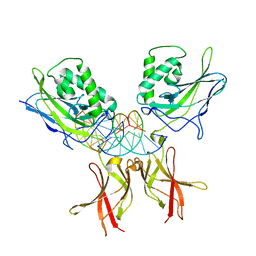 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with 10-mer kappaB DNA from human Neutrophil Gelatinase-associated Lipocalin (NGAL) promoter | | Descriptor: | DNA A, DNA B, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Zhu, N, Mealka, M, Mitchel, S, Rogers, W.E, Huxford, T. | | Deposit date: | 2023-07-25 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
8TKM
 
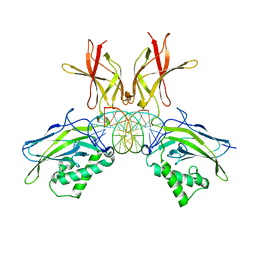 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with 17-mer kappaB DNA from human interleukin-6 (IL-6) promoter | | Descriptor: | 17-mer kappaB DNA, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Zhu, N, Mealka, M, Mitchel, S, Rogers, W.E, Huxford, T. | | Deposit date: | 2023-07-25 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
6JUZ
 
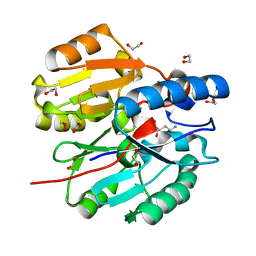 | | Crystal Structure of N-terminal domain of ArgZ(N71S) covalently bond to a reaction intermediate | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhuang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
6JUY
 
 | | Crystal Structure of ArgZ, apo structure, an Arginine Dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
5JK5
 
 | |
5JK6
 
 | |
5JK8
 
 | |
6JV0
 
 | | Crystal Structure of N-terminal domain of ArgZ, bound to Product, an arginine dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | 1,2-ETHANEDIOL, L-ornithine, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
6JV1
 
 | | Crystal Structure of N-terminal domain of ArgZ, C264S mutant, bound to Substrate, an arginine dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | ARGININE, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
4HU0
 
 | |
4HTY
 
 | |
4HAC
 
 | |
9BOR
 
 | | IkappaBzeta ankyrin repeat domain:NF-kappaB p50 homodimer complex at 2.0 Angstrom resolution | | Descriptor: | (2R,3S)-heptane-1,2,3-triol, NF-kappa-B inhibitor zeta, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Rogers, W.E, Zhu, N, Huxford, T. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural and biochemical analyses of the nuclear I kappa B zeta protein in complex with the NF-kappa B p50 homodimer.
Genes Dev., 38, 2024
|
|
3UK7
 
 | |
2H5U
 
 | | Crystal structure of laccase from Cerrena maxima at 1.9A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lyashenko, A.V, Gabdoulkhakov, A.G, Zaitsev, V.N, Lamzin, V.S, Lindley, P.F, Bento, I, Betzel, C, Zhukhlistova, N.E, Zhukova, Y.N, Mikhailov, A.M. | | Deposit date: | 2006-05-27 | | Release date: | 2007-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Purification, crystallization and preliminary X-ray study of the fungal laccase from Cerrena maxima
Acta Crystallogr.,Sect.F, 62, 2006
|
|
4DML
 
 | |
4DMM
 
 | |
3ORF
 
 | | Crystal Structure of Dihydropteridine Reductase from Dictyostelium discoideum | | Descriptor: | Dihydropteridine reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chen, C, Zhuang, N.N, Seo, K.H, Park, Y.S, Lee, K.H. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural insights into the dual substrate specificities of mammalian and Dictyostelium dihydropteridine reductases toward two stereoisomers of quinonoid dihydrobiopterin
Febs Lett., 585, 2011
|
|
1SK6
 
 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin, 3',5' cyclic AMP (cAMP), and pyrophosphate | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CALCIUM ION, Calmodulin, ... | | Authors: | Guo, Q, Shen, Y, Zhukovskaya, N.L, Tang, W.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and kinetic analyses of the interaction of anthrax adenylyl cyclase toxin with reaction products cAMP and pyrophosphate.
J.Biol.Chem., 279, 2004
|
|
1XFX
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin in the presence of 10 millimolar exogenously added calcium chloride | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XFZ
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin in the presence of 1 millimolar exogenously added calcium chloride | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
8UPS
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 5 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, PHOSPHATE ION | | Authors: | Wu, Y, Qiang, D, Zhuang, N, Krishnamurthy, H, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
1Y0V
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and pyrophosphate | | Descriptor: | CALCIUM ION, Calmodulin, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.-J. | | Deposit date: | 2004-11-16 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor
Embo J., 24, 2005
|
|
1XFU
 
 | | Crystal structure of anthrax edema factor (EF) truncation mutant, EF-delta 64 in complex with calmodulin | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XFY
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
