5YPO
 
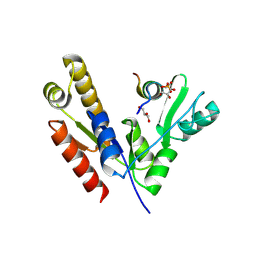 | | Crystal structure of PSD-95 GK domain in complex with phospho-SAPAP peptide | | Descriptor: | Disks large homolog 4, GLYCEROL, SAPAP | | Authors: | Zhu, J, Zhou, Q, Shang, Y, Weng, Z, Zhang, R, Zhang, M. | | Deposit date: | 2017-11-02 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Synaptic Targeting and Function of SAPAPs Mediated by Phosphorylation-Dependent Binding to PSD-95 MAGUKs.
Cell Rep, 21, 2017
|
|
6JMT
 
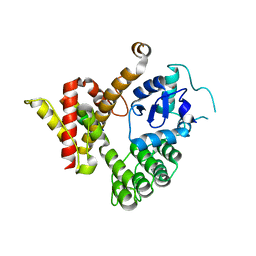 | | Crystal structure of GIT/PIX complex | | Descriptor: | ARF GTPase-activating protein GIT2, ZINC ION, beta PIX | | Authors: | Zhu, J, Lin, L, Xia, Y, Zhang, R, Zhang, M. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling.
Mol.Cell, 79, 2020
|
|
6JMU
 
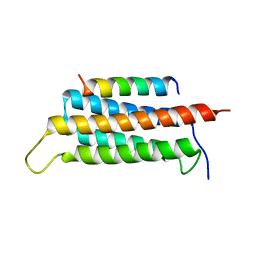 | | Crystal structure of GIT1/Paxillin complex | | Descriptor: | ARF GTPase-activating protein GIT1, Paxillin | | Authors: | Zhu, J, Lin, L, Xia, Y, Zhang, R, Zhang, M. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling.
Mol.Cell, 79, 2020
|
|
2KBK
 
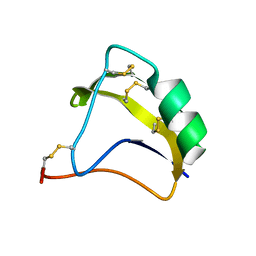 | | Solution Structure of BmK-M10 | | Descriptor: | Neurotoxin BmK-M10 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-22 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmK-M10
To be Published
|
|
2KBH
 
 | | solution structure of BmKalphaTx11 (major conformation) | | Descriptor: | Toxin Bmka2 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKalphaTx11, a toxin from the venom of the Chinese scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 391, 2010
|
|
2KBJ
 
 | | solution structure of BmKalphaTx11 (minor conformation) | | Descriptor: | Toxin Bmka2 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKalphaTx11, a toxin from the venom of the Chinese scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 391, 2010
|
|
7E85
 
 | | SnoaL-like domain-containing protein | | Descriptor: | SnoaL-like domain-containing protein | | Authors: | Zhuang, J. | | Deposit date: | 2021-03-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | SnoaL-like domain-containing protein
To Be Published
|
|
4V63
 
 | | Structural basis for translation termination on the 70S ribosome. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Laurberg, M, Asahara, H, Korostelev, A, Zhu, J, Trakhanov, S, Noller, H.F. | | Deposit date: | 2008-05-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | Structural basis for translation termination on the 70S ribosome
Nature, 454, 2008
|
|
4V7P
 
 | | Recognition of the amber stop codon by release factor RF1. | | Descriptor: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Zhu, J, Asahara, H, Noller, H.F. | | Deposit date: | 2010-04-29 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Recognition of the amber UAG stop codon by release factor RF1.
Embo J., 29, 2010
|
|
4V9N
 
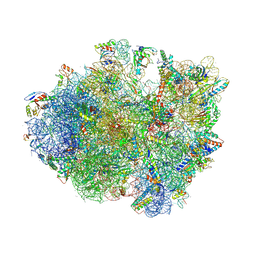 | | Crystal structure of the 70S ribosome bound with the Q253P mutant of release factor RF2. | | Descriptor: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Santos, N, Zhu, J, Donohue, J.P, Korostelev, A.A, Noller, H.F. | | Deposit date: | 2013-04-26 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of the 70S Ribosome Bound with the Q253P Mutant Form of Release Factor RF2.
Structure, 21, 2013
|
|
1T4J
 
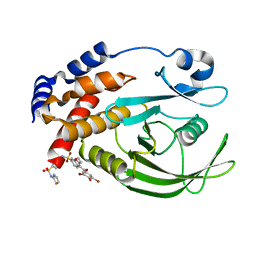 | | Allosteric Inhibition of Protein Tyrosine Phosphatase 1B | | Descriptor: | 3-(3,5-DIBROMO-4-HYDROXY-BENZOYL)-2-ETHYL-BENZOFURAN-6-SULFONIC ACID [4-(THIAZOL-2-YLSULFAMOYL)-PHENYL]-AMIDE, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Wiesmann, C, Barr, K.J, Kung, J, Zhu, J, Shen, W, Fahr, B.J, Zhong, M, Taylor, L, Randal, M, McDowell, R.S, Hansen, S.K. | | Deposit date: | 2004-04-29 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric inhibition of protein tyrosine phosphatase 1B
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T48
 
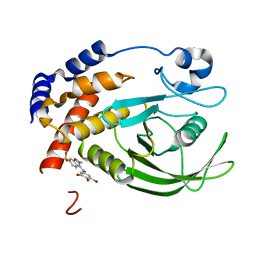 | | Allosteric Inhibition of Protein Tyrosine Phosphatase 1B | | Descriptor: | 3-(3,5-DIBROMO-4-HYDROXY-BENZOYL)-2-ETHYL-BENZOFURAN-6-SULFONIC ACID DIMETHYLAMIDE, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Wiesmann, C, Barr, K.J, Kung, J, Zhu, J, Shen, W, Fahr, B.J, Zhong, M, Erlanson, D.A, Taylor, L, Randal, M, McDowell, R.S, Hansen, S.K. | | Deposit date: | 2004-04-28 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric inhibition of protein tyrosine phosphatase 1B
Nat.Struct.Mol.Biol., 11, 2004
|
|
6CKB
 
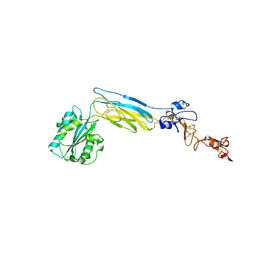 | | Crystal structure of an extended beta3 integrin P33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Chimera protein of Integrin beta-3 and Integrin alpha-L, ... | | Authors: | Zhou, D, Zhu, J. | | Deposit date: | 2018-02-27 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an extended beta3integrin.
Blood, 132, 2018
|
|
4RR4
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A367 | | Descriptor: | 2-chloro-N-[3-(4-{[(2Z)-2-cyano-3-cyclopropyl-3-hydroxyprop-2-enoyl]amino}phenoxy)phenyl]-4-methyl-1,3-thiazole-5-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-11-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A367
TO BE PUBLISHED
|
|
4RKA
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A347 | | Descriptor: | 2-{[5-(naphthalen-1-ylmethyl)-4-oxo-4H-1lambda~4~,3-thiazol-2-yl]amino}benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-10-12 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A347
TO BE PUBLISHED
|
|
4RLI
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A048 | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-10-17 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A048
TO BE PUBLISHED
|
|
4RK8
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A356 | | Descriptor: | 5-fluoro-2-{[(5Z)-5-(naphthalen-1-ylmethylidene)-4-oxo-4,5-dihydro-1,3-thiazol-2-yl]amino}benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-10-12 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A356
TO BE PUBLISHED
|
|
4UQ8
 
 | | Electron cryo-microscopy of bovine Complex I | | Descriptor: | ACYL CARRIER PROTEIN, MITOCHONDRIAL, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2014-06-21 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.95 Å) | | Cite: | Architecture of Mammalian Respiratory Complex I.
Nature, 515, 2014
|
|
6PPA
 
 | | Crystal structure of the unliganded bromodomain of human BRD7 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 7, PHOSPHATE ION | | Authors: | Chan, A, Karim, M.R, Zhu, J, Schonbrunn, E. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6IF3
 
 | | Complex structure of Rab35 and its effector ACAP2 | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lin, L, Zhu, J, Zhang, R. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rab35/ACAP2 and Rab35/RUSC2 Complex Structures Reveal Molecular Basis for Effector Recognition by Rab35 GTPase.
Structure, 27, 2019
|
|
6JIQ
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6IF2
 
 | | Complex structure of Rab35 and its effector RUSC2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Iporin, MAGNESIUM ION, ... | | Authors: | Lin, L, Zhu, J, Zhang, R. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rab35/ACAP2 and Rab35/RUSC2 Complex Structures Reveal Molecular Basis for Effector Recognition by Rab35 GTPase.
Structure, 27, 2019
|
|
8HWD
 
 | | Cryo-EM Structure of D5 ADP form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Primase D5 | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWA
 
 | | D5 ATP-ADP-Apo-ssDNA IS1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWG
 
 | | D5 ATPrS-ADP-ssDNA form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
