6PM4
 
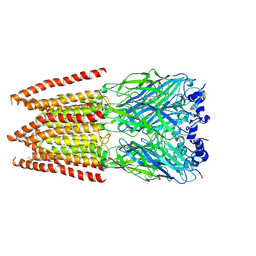 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Glycine in SMA, super-open state | | Descriptor: | GLYCINE, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM2
 
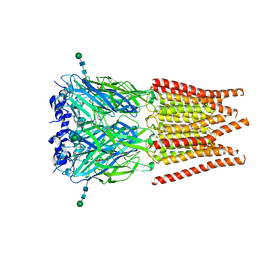 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Taurine in SMA, open state | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM6
 
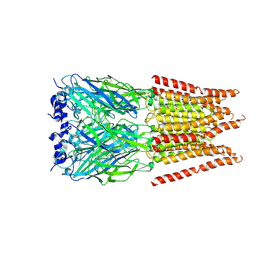 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Glycine in SMA, open state | | Descriptor: | GLYCINE, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLW
 
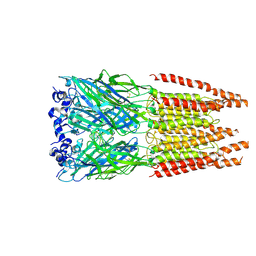 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in SMA, super-open state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6QF8
 
 | |
6QH2
 
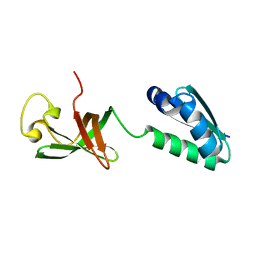 | |
6QFP
 
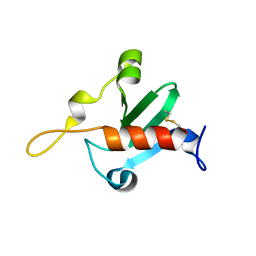 | |
8ISZ
 
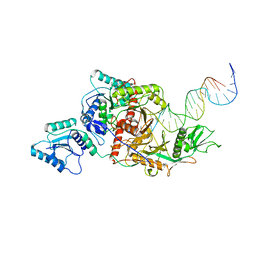 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA monomer | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISY
 
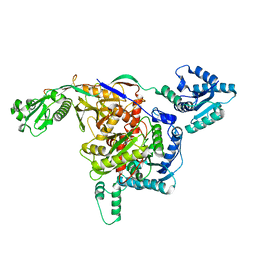 | | Cryo-EM structure of free-state Crt-SPARTA | | Descriptor: | Piwi domain-containing protein, TIR domain-containing protein | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
5FZQ
 
 | | Designed TPR Protein M4N | | Descriptor: | DESIGNED TPR PROTEIN, SULFATE ION | | Authors: | Albrecht, R, Zhu, H, Hartmann, M.D. | | Deposit date: | 2016-03-15 | | Release date: | 2016-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Origin of a folded repeat protein from an intrinsically disordered ancestor.
Elife, 5, 2016
|
|
5FZR
 
 | |
5DTH
 
 | |
4IQ6
 
 | | Gsk-3beta with inhibitor 6-chloro-N-cyclohexyl-4-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyridin-2-amine | | Descriptor: | 6-chloro-N-cyclohexyl-4-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyridin-2-amine, Glycogen synthase kinase-3 beta | | Authors: | Tong, Y, Stewart, K.D, Florjancic, A.S, Harlan, J.E, Merta, P.J, Przytulinska, M, Soni, N, Swinger, K.S, Zhu, H, Johnson, E.F, Shoemaker, A.R, Penning, T.D. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Azaindole-Based Inhibitors of Cdc7 Kinase: Impact of the Pre-DFG Residue, Val 195.
ACS Med Chem Lett, 4, 2013
|
|
6N5V
 
 | | Crystal Structure of Strictosidine in complex with 1H-indole-4-ethanamine | | Descriptor: | 2-(1H-indol-4-yl)ethan-1-amine, Strictosidine synthase | | Authors: | Cai, Y, Shao, N, Xie, H, Futamura, Y, Panjikar, S, Liu, H, Zhu, H, Osada, H, Zou, H. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Crystal Structure of Strictosidine in complex with 1H-indole-4-ethanamine
to be published
|
|
4JLW
 
 | | Crystal structure of formaldehyde dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | Glutathione-independent formaldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Chen, S, Liao, Y.P, Wang, D.L, Wang, S, Ding, J.F, Wang, Y.M, Cai, L.J, Ran, X.Y, Zhu, H.X. | | Deposit date: | 2013-03-13 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of formaldehyde dehydrogenase from Pseudomonas aeruginosa: the binary complex with the cofactor NAD+.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5FZS
 
 | |
5HSI
 
 | | Crystal structure of tyrosine decarboxylase at 1.73 Angstroms resolution | | Descriptor: | MAGNESIUM ION, Putative decarboxylase | | Authors: | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | Deposit date: | 2016-01-25 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
6NJE
 
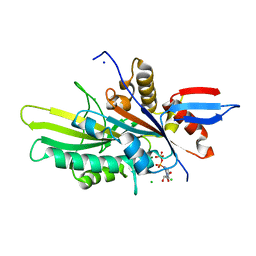 | | Crystal structure of the motor domain of human kinesin family member 22 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Kinesin-like protein KIF22, ... | | Authors: | Walker, B.C, Zhu, H, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Park, H, Cochran, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-03 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the motor domain of human kinesin family member 22
To Be Published
|
|
8W4F
 
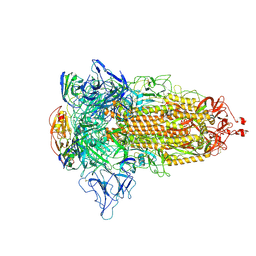 | | SARS-CoV-2 spike protein in complex with a trivalent nanobody | | Descriptor: | Spike glycoprotein, Tribody | | Authors: | Jiang, X.Y, Qian, J.Q, Zhu, H.X, Qin, Q, Huang, Q. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure-guided design of a trivalent nanobody cluster targeting SARS-CoV-2 spike protein.
Int.J.Biol.Macromol., 256, 2024
|
|
6DW0
 
 | | Cryo-EM structure of the benzodiazepine-sensitive alpha1beta1gamma2S tri-heteromeric GABAA receptor in complex with GABA (Whole map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1,Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Phulera, S, Zhu, H, Yu, J, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the benzodiazepine-sensitive alpha 1 beta 1 gamma 2S tri-heteromeric GABAAreceptor in complex with GABA.
Elife, 7, 2018
|
|
6DW1
 
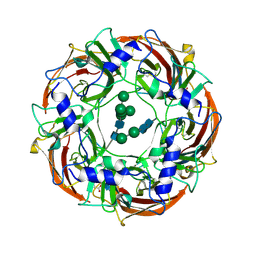 | | Cryo-EM structure of the benzodiazepine-sensitive alpha1beta1gamma2S tri-heteromeric GABAA receptor in complex with GABA (ECD map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1,Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Phulera, S, Zhu, H, Yu, J, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the benzodiazepine-sensitive alpha 1 beta 1 gamma 2S tri-heteromeric GABAAreceptor in complex with GABA.
Elife, 7, 2018
|
|
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | Descriptor: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
6MV1
 
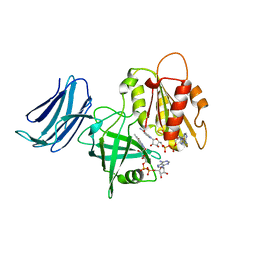 | | 2.15A resolution structure of the CS-b5R domains of human Ncb5or (NAD+ form) | | Descriptor: | Cytochrome b5 reductase 4, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Benson, D.R, Cooper, A, Gao, P, Zhu, H. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of the naturally fused CS and cytochrome b5reductase (b5R) domains of Ncb5or reveal an expanded CS fold, extensive CS-b5R interactions and productive binding of the NAD(P)+nicotinamide ring.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6MV2
 
 | | 2.05A resolution structure of the CS-b5R domains of human Ncb5or (NADP+ form) | | Descriptor: | Cytochrome b5 reductase 4, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Benson, D.R, Cooper, A, Gao, P, Zhu, H. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the naturally fused CS and cytochrome b5reductase (b5R) domains of Ncb5or reveal an expanded CS fold, extensive CS-b5R interactions and productive binding of the NAD(P)+nicotinamide ring.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5U8A
 
 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(2-bromo-6-fluorophenyl)methyl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(2-bromo-6-fluorophenyl)methyl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
