1X7I
 
 | | Crystal structure of the native copper homeostasis protein (cutCm) with calcium binding from Shigella flexneri 2a str. 301 | | Descriptor: | CALCIUM ION, Copper homeostasis protein cutC | | Authors: | Zhu, D.Y, Zhu, Y.Q, Huang, R.H, Xiang, Y, Wang, D.C. | | Deposit date: | 2004-08-14 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the copper homeostasis protein (CutCm) from Shigella flexneri at 1.7 A resolution: The first structure of a new sequence family of TIM barrels
Proteins, 58, 2004
|
|
4U03
 
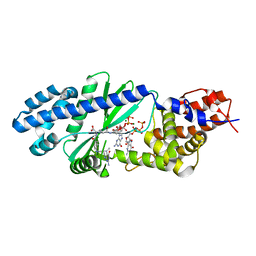 | |
2F5I
 
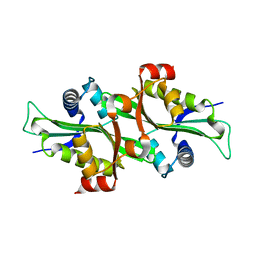 | |
1X8C
 
 | |
2DS2
 
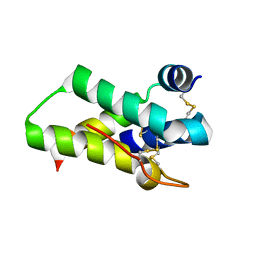 | | Crystal structure of mabinlin II | | Descriptor: | ACETIC ACID, Sweet protein mabinlin-2 chain A, Sweet protein mabinlin-2 chain B | | Authors: | Li, D.F, Zhu, D.Y, Wang, D.C. | | Deposit date: | 2006-06-19 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Mabinlin II: a novel structural type of sweet proteins and the main structural basis for its sweetness.
J.Struct.Biol., 162, 2008
|
|
2ZJT
 
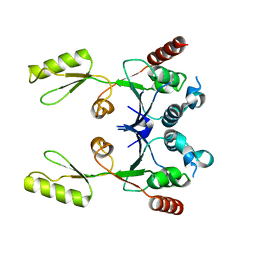 | |
4U0L
 
 | |
4U0N
 
 | | Structure of the Vibrio cholerae di-nucleotide cyclase (DncV) deletion mutant D-loop | | Descriptor: | Cyclic AMP-GMP synthase, MAGNESIUM ION, N-[4-({[(6S)-2-amino-5-methyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-gamma-glutamyl-L-glutamic acid | | Authors: | Xiang, Y, Zhu, D.Y. | | Deposit date: | 2014-07-12 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural Biochemistry of a Vibrio cholerae Dinucleotide Cyclase Reveals Cyclase Activity Regulation by Folates.
Mol.Cell, 55, 2014
|
|
2VHA
 
 | | DEBP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTAMIC ACID, PERIPLASMIC BINDING TRANSPORT PROTEIN | | Authors: | Hu, Y.L, Fan, C.-P, Fu, G.S, Zhu, D.Y, Jin, Q, Wang, D.-C. | | Deposit date: | 2007-11-20 | | Release date: | 2008-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of a Glutamate/Aspartate Binding Protein Complexed with a Glutamate Molecule: Structural Basis of Ligand Specificity at Atomic Resolution.
J.Mol.Biol., 382, 2008
|
|
5Y66
 
 | | Crystal structure of Pseudomonas fluorescens Kynurenine 3-monooxygenase in complex with L-KYN and Ro61-8048 | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 3,4-dimethoxy-N-[4-(3-nitrophenyl)-1,3-thiazol-2-yl]benzenesulfonamide, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xiang, Y, Gao, J.J, Zhu, D.Y. | | Deposit date: | 2017-08-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Biochemistry and structural studies of kynurenine 3-monooxygenase reveal allosteric inhibition by Ro 61-8048
FASEB J., 32, 2018
|
|
5Y7A
 
 | |
5Y77
 
 | |
