7UOL
 
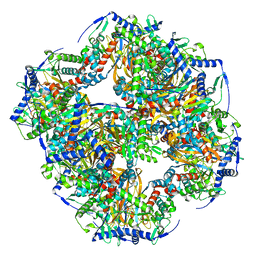 | | Endogenous dihydrolipoamide succinyltransferase (E2) core of 2-oxoglutarate dehydrogenase complex from bovine kidney | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | | Authors: | Liu, S, Xia, X, Zhen, J, Li, Z.H, Zhou, Z.H. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and comparison of endogenous 2-oxoglutarate and pyruvate dehydrogenase complexes from bovine kidney.
Cell Discov, 8, 2022
|
|
7UOM
 
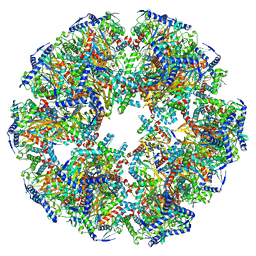 | | Endogenous dihydrolipoamide acetyltransferase (E2) core of pyruvate dehydrogenase complex from bovine kidney | | Descriptor: | Acetyltransferase component of pyruvate dehydrogenase complex | | Authors: | Liu, S, Xia, X, Zhen, J, Li, Z.H, Zhou, Z.H. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures and comparison of endogenous 2-oxoglutarate and pyruvate dehydrogenase complexes from bovine kidney.
Cell Discov, 8, 2022
|
|
1CAD
 
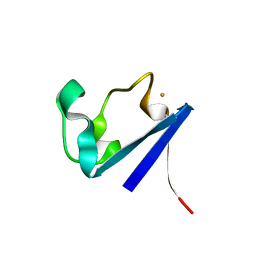 | | X-RAY CRYSTAL STRUCTURES OF THE OXIDIZED AND REDUCED FORMS OF THE RUBREDOXIN FROM THE MARINE HYPERTHERMOPHILIC ARCHAEBACTERIUM PYROCOCCUS FURIOSUS | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Day, M.W, Hsu, B.T, Joshua-Tor, L, Park, J.B, Zhou, Z.H, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1992-05-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus.
Protein Sci., 1, 1992
|
|
1CAA
 
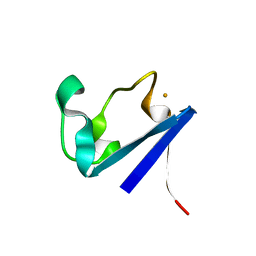 | | X-RAY CRYSTAL STRUCTURES OF THE OXIDIZED AND REDUCED FORMS OF THE RUBREDOXIN FROM THE MARINE HYPERTHERMOPHILIC ARCHAEBACTERIUM PYROCOCCUS FURIOSUS | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Day, M.W, Hsu, B.T, Joshua-Tor, L, Park, J.B, Zhou, Z.H, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1992-05-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus.
Protein Sci., 1, 1992
|
|
7SLQ
 
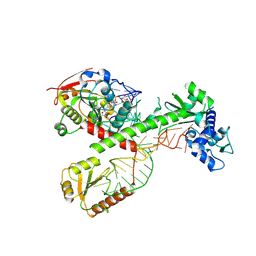 | | Cryo-EM structure of 7SK core RNP with circular RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, La-related protein 7, Minimal circular 7SK RNA, ... | | Authors: | Yang, Y, Liu, S, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-10-24 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP.
Mol.Cell, 82, 2022
|
|
7SLP
 
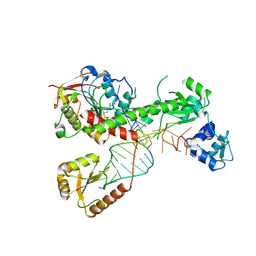 | | Cryo-EM structure of 7SK core RNP with linear RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, La-related protein 7, Linear 7SK RNA, ... | | Authors: | Yang, Y, Liu, S, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-10-24 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP.
Mol.Cell, 82, 2022
|
|
7TW5
 
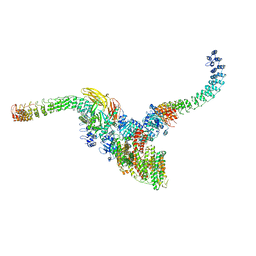 | | Cryo-EM structure of human ankyrin complex (B2P1A2) from red blood cell | | Descriptor: | Ankyrin-1, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TW6
 
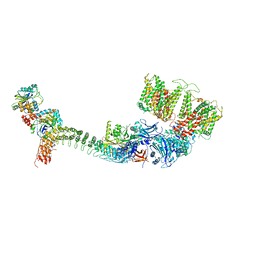 | | Cryo-EM structure of human ankyrin complex (B4P1A1) from red blood cell | | Descriptor: | Ankyrin-1, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TVZ
 
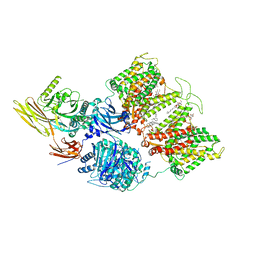 | | Cryo-EM structure of human band 3-protein 4.2 complex in diagonal conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CHOLESTEROL, ... | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TW3
 
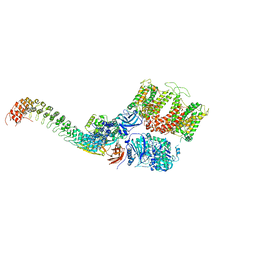 | | Cryo-EM structure of human ankyrin complex (B2P1A1) from red blood cell | | Descriptor: | Ankyrin-1, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TW2
 
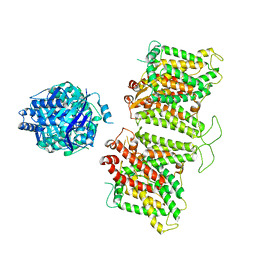 | |
7TW1
 
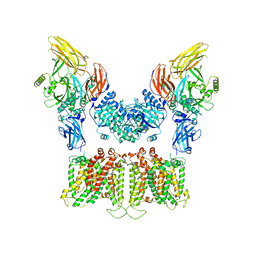 | |
7TW0
 
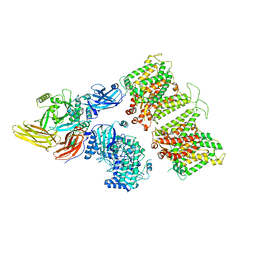 | | Cryo-EM structure of human band 3-protein 4.2 complex in vertical conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6U5J
 
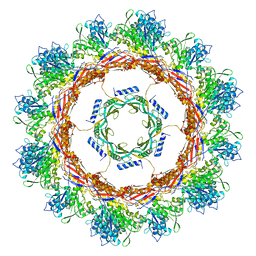 | | CryoEM Structure of Pyocin R2 - postcontracted - collar | | Descriptor: | Collar PA0615, Sheath PA0622 | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
6UUR
 
 | | Human prion protein fibril, M129 variant | | Descriptor: | Major prion protein | | Authors: | Glynn, C, Sawaya, M.R, Ge, P, Zhou, Z.H, Rodriguez, J.A. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of a human prion fibril with a hydrophobic, protease-resistant core.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8F41
 
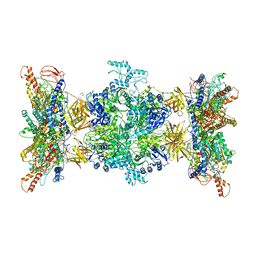 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
8FJP
 
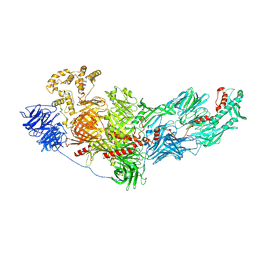 | | Cryo-EM structure of native mosquito salivary gland surface protein 1 (SGS1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, salivary gland surface protein 1 | | Authors: | Liu, S, Xia, X, Calvo, E, Zhou, Z.H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Native structure of mosquito salivary protein uncovers domains relevant to pathogen transmission.
Nat Commun, 14, 2023
|
|
8FJL
 
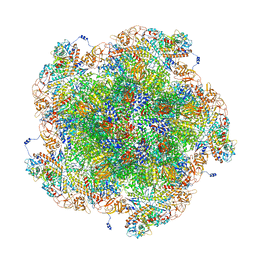 | | Golden Shiner Reovirus Core Tropical Vertex | | Descriptor: | Clamp protein VP6, Major inner capsid protein VP3, Microtubule-associated protein VP5, ... | | Authors: | Stevens, A.S, Zhou, Z.H. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Asymmetric reconstruction of the aquareovirus core at near-atomic resolution and mechanism of transcription initiation.
Protein Cell, 14, 2023
|
|
8FJK
 
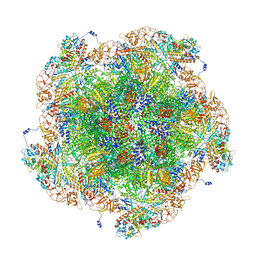 | | Golden Shiner Reovirus Core Polar Vertex | | Descriptor: | Clamp protein VP6, Major inner capsid protein VP3, Microtubule-associated protein VP5, ... | | Authors: | Stevens, A.S, Zhou, Z.H. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Asymmetric reconstruction of the aquareovirus core at near-atomic resolution and mechanism of transcription initiation.
Protein Cell, 14, 2023
|
|
6VOC
 
 | | icosahedral symmetry reconstruction of brome mosaic virus (RNA 3+4) | | Descriptor: | Capsid protein | | Authors: | Beren, C, Cui, Y.X, Chakravarty, A, Yang, X, Rao, A.L.N, Knobler, C.M, Zhou, Z.H, Gelbart, W.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Genome organization and interaction with capsid protein in a multipartite RNA virus.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8FN4
 
 | | Cryo-EM structure of RNase-treated RESC-A in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), RNA-editing substrate-binding complex protein 3 (RESC3), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN6
 
 | | Cryo-EM structure of RNase-untreated RESC-A in trypanosomal RNA editing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
