7V61
 
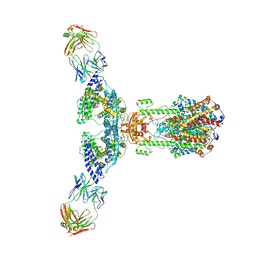 | | ACE2 -Targeting Monoclonal Antibody as Potent and Broad-Spectrum Coronavirus Blocker | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3E8, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2021-08-18 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | ACE2-targeting monoclonal antibody as potent and broad-spectrum coronavirus blocker.
Signal Transduct Target Ther, 6, 2021
|
|
7WWM
 
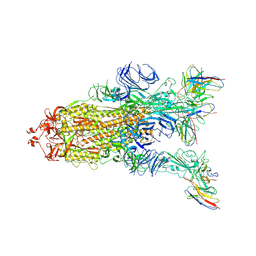 | | S protein of Delta variant in complex with ZWC6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-13 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination.
Signal Transduct Target Ther, 7, 2022
|
|
7WWL
 
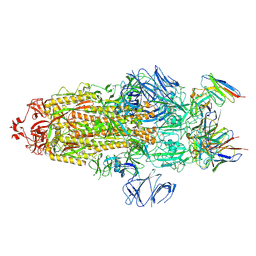 | | S protein of Delta variant in complex with ZWD12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-13 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination.
Signal Transduct Target Ther, 7, 2022
|
|
7XLM
 
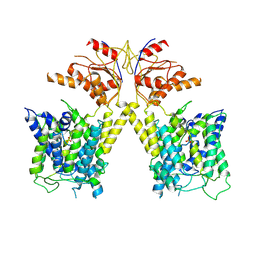 | |
7XUL
 
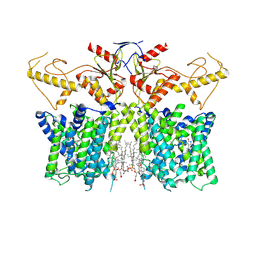 | | Human SLC26A3 in complex with Tenidap | | Descriptor: | 5-chloranyl-2-oxidanyl-3-thiophen-2-ylcarbonyl-indole-1-carboxamide, CHOLESTEROL HEMISUCCINATE, Chloride anion exchanger | | Authors: | Li, X.R, Chi, X.M, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of human SLC26A3 in complex with Tenidap
To Be Published
|
|
7XUJ
 
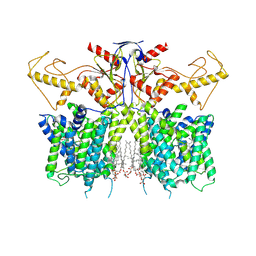 | | Human SLC26A3 in complex with UK5099 | | Descriptor: | (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid, CHOLESTEROL HEMISUCCINATE, Chloride anion exchanger | | Authors: | Li, X.R, Chi, X.M, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | human SLC26A3 in complex with UK5099
To Be Published
|
|
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
7XUH
 
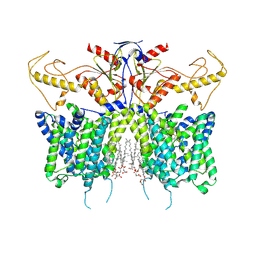 | | Down-regulated in adenoma in complex with TQR1122 | | Descriptor: | 2-[4,8-dimethyl-2-oxidanylidene-7-[[3-(trifluoromethyl)phenyl]methoxy]chromen-3-yl]ethanoic acid, CHOLESTEROL HEMISUCCINATE, Chloride anion exchanger | | Authors: | Li, X.R, Chi, X.M, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | structure of human SLC26A3 in complex with TQR1122
To Be Published
|
|
7WWB
 
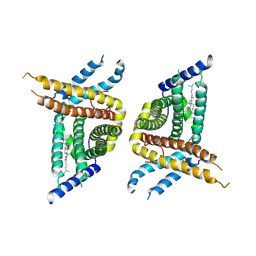 | | Choline transporter-like protein 1 | | Descriptor: | CHOLESTEROL, CHOLINE ION, Choline transporter-like protein 1 | | Authors: | Chi, X.M, Zhou, Q. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Rational exploration of fold atlas for human solute carrier proteins.
Structure, 30, 2022
|
|
7VTV
 
 | | Crystal structure of PDE8A catalytic domain in complex with 15 | | Descriptor: | 2-chloro-9-(3-(2,2-difluoroethoxy)-5-(difluoromethoxy)benzyl)-9H-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wu, X.-N, Zhou, Q, Huang, Y.-D, Li, Z, Wu, Y, Luo, H.-B. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Discovery of Orally Efficient PDE8 Inhibitors for the Treatment of Vascular Dementia
To Be Published
|
|
7VTX
 
 | | Crystal structure of PDE8A catalytic domain in complex with 22 | | Descriptor: | 2-chloro-9-(3-(2,2-difluoroethoxy)-5-(pyridin-4-yl)benzyl)-9H-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wu, X.-N, Zhou, Q, Huang, Y.-D, Li, Z, Wu, Y, Luo, H.-B. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.50010943 Å) | | Cite: | Structure-Based Discovery of Orally Efficient PDE8 Inhibitors for the Treatment of Vascular Dementia
To Be Published
|
|
7VSL
 
 | | Crystal structure of PDE8A catalytic domain in complex with 10 | | Descriptor: | 2-chloro-9-(3-(2,2-difluoroethoxy)benzyl)-9H-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wu, X.-N, Zhou, Q, Huang, Y.-D, Li, Z, Wu, Y, Luo, H.-B. | | Deposit date: | 2021-10-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.500069 Å) | | Cite: | Structure-Based Discovery of Orally Efficient PDE8 Inhibitors for the Treatment of Vascular Dementia
To Be Published
|
|
7VTW
 
 | | Crystal structure of PDE8A catalytic domain in complex with 17 | | Descriptor: | 2-chloro-9-(3-(2,2-difluoroethoxy)-5-isopropoxybenzyl)-9H-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wu, X.-N, Zhou, Q, Huang, Y.-D, Li, Z, Wu, Y, Luo, H.-B. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79971337 Å) | | Cite: | Structure-Based Discovery of Orally Efficient PDE8 Inhibitors for the Treatment of Vascular Dementia
To Be Published
|
|
4CMH
 
 | | Crystal structure of CD38 with a novel CD38-targeting antibody SAR650984 | | Descriptor: | ADP-RIBOSYL CYCLASE 1, HEAVY CHAIN OF SAR650984-FAB FRAGMENT, LIGHT CHAIN OF SAR650984-FAB FRAGMENT | | Authors: | Deckert, J, Wetzel, M.C, Park, P.U, Bartle, L.M, Skaletskaya, A, Goldmacher, V, Vallee, F, ZhouLiu, Q, Ferrari, P, Pouzieux, S, Lahoute, C, Dumontet, C, Plesa, A, Chiron, M, Lejeune, P, Chittenden, T, Blanc, V. | | Deposit date: | 2014-01-15 | | Release date: | 2014-07-16 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | SAR650984, a novel humanized CD38-targeting antibody, demonstrates potent antitumor activity in models of multiple myeloma and other CD38+ hematologic malignancies.
Clin. Cancer Res., 20, 2014
|
|
