6LR8
 
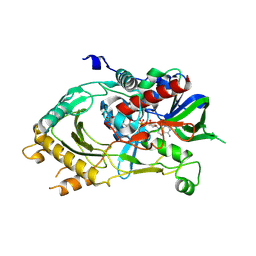 | |
7BWP
 
 | | Crystal complex of endo-deglycosylated PcHNL5 with (R)-mandelonitrile | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zheng, Y.C, Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure-Guided Tuning of a Hydroxynitrile Lyase to Accept Rigid Pharmaco Aldehydes.
Acs Catalysis, 2020
|
|
6LQY
 
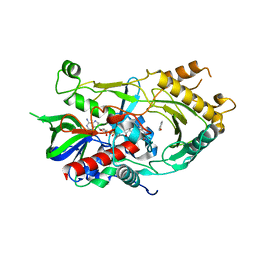 | | Crystal complex of endo-deglycosylated hydroxynitrile lyase isozyme 5 of Prunus communis with benzaldehyde | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PREDICTED: (R)-mandelonitrile lyase, ... | | Authors: | Zheng, Y.C, Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2020-01-15 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Structure-Guided Tuning of a Hydroxynitrile Lyase to Accept Rigid Pharmaco Aldehydes.
Acs Catalysis, 2020
|
|
6WY6
 
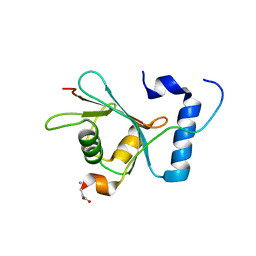 | | Crystal structure of S. cerevisiae Atg8 in complex with Ede1 (1220-1247) | | Descriptor: | Autophagy-related protein 8, EH domain-containing and endocytosis protein 1 | | Authors: | Zheng, Y, Wilfling, F, Baumeister, W, Schulman, B.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | A Selective Autophagy Pathway for Phase-Separated Endocytic Protein Deposits.
Mol.Cell, 80, 2020
|
|
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
7WXZ
 
 | | Crystal structure of the recombinant protein HR121 from the S2 protein of SARS-CoV-2 | | Descriptor: | Spike protein S2' | | Authors: | Zheng, Y.T, Ouyang, S, Pang, W, Lu, Y, Zhao, Y.B. | | Deposit date: | 2022-02-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A variant-proof SARS-CoV-2 vaccine targeting HR1 domain in S2 subunit of spike protein.
Cell Res., 32, 2022
|
|
7S5I
 
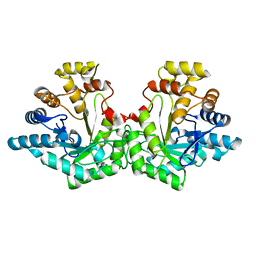 | | Crystal structure of Aldose-6-phosphate reductase (Ald6PRase) from peach (Prunus persica) leaves | | Descriptor: | Sorbitol-6-phosphate dehydrogenase | | Authors: | Zheng, Y, Bhayani, J.A, Romina, I.M, Hartman, M.D, Cereijo, A.E, Ballicora, M.A, Iglesias, A.A, Figueroa, C.M, Liu, D. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural Determinants of Sugar Alcohol Biosynthesis in Plants: The Crystal Structures of Mannose-6-Phosphate and Aldose-6-Phosphate Reductases.
Plant Cell.Physiol., 63, 2022
|
|
1G72
 
 | | CATALYTIC MECHANISM OF QUINOPROTEIN METHANOL DEHYDROGENASE: A THEORETICAL AND X-RAY CRYSTALLOGRAPHIC INVESTIGATION | | Descriptor: | CALCIUM ION, METHANOL DEHYDROGENASE HEAVY SUBUNIT, METHANOL DEHYDROGENASE LIGHT SUBUNIT, ... | | Authors: | Zheng, Y, Xia, Z, Chen, Z, Bruice, T.C, Mathews, F.S. | | Deposit date: | 2000-11-08 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic mechanism of quinoprotein methanol dehydrogenase: A theoretical and x-ray crystallographic investigation.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3WGH
 
 | | Crystal structure of RSP in complex with beta-NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CACODYLATE ION, Redox-sensing transcriptional repressor rex, ... | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
3WGI
 
 | | Crystal structure of RSP in complex with beta-NAD+ and operator DNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*TP*GP*TP*TP*AP*AP*TP*CP*GP*AP*TP*TP*AP*AP*CP*AP*AP*TP*C)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), Redox-sensing transcriptional repressor rex | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
3WG9
 
 | | Crystal structure of RSP, a Rex-family repressor | | Descriptor: | Redox-sensing transcriptional repressor rex, SULFATE ION | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-03 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
7S5F
 
 | | Crystal structure of mannose-6-phosphate reductase from celery (Apium graveolens) leaves with NADP+ and mannonic acid bound | | Descriptor: | D-MANNONIC ACID, Manose-6-phosphate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, Y, Bhayani, J.A, Romina, I.M, Hartman, M.D, Cereijo, A.E, Ballicora, M.A, Iglesias, A.A, Figueroa, C.M, Liu, D. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Determinants of Sugar Alcohol Biosynthesis in Plants: The Crystal Structures of Mannose-6-Phosphate and Aldose-6-Phosphate Reductases.
Plant Cell.Physiol., 63, 2022
|
|
5IE7
 
 | | Crystal structure of a lactonase double mutant in complex with substrate b | | Descriptor: | (3S,7S,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | Authors: | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
5IE6
 
 | | Crystal structure of a lactonase mutant in complex with substrate b | | Descriptor: | (3S,7S,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | Authors: | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
5IE4
 
 | | Crystal structure of a lactonase mutant in complex with substrate a | | Descriptor: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | Authors: | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
5IE5
 
 | | Crystal structure of a lactonase double mutant in complex with substrate a | | Descriptor: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | Authors: | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
8JM5
 
 | | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A/S333V/P340L from Prunus communis | | Descriptor: | (R)-mandelonitrile lyase, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Y.-C, Li, F.-L, Yu, H.-L, Xu, J.-H. | | Deposit date: | 2023-06-04 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A/S333V/P340L from Prunus communis
To Be Published
|
|
8JM6
 
 | | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A/S333V/P340L from Prunus communis complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde (catalytic conformation) | | Descriptor: | (R)-mandelonitrile lyase, 2,2-dimethyl-4H-1,3-benzodioxine-6-carbaldehyde, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Y.-C, Li, F.-L, Yu, H.-L, Xu, J.-H. | | Deposit date: | 2023-06-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A/S333V/P340L from Prunus communis complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde (catalytic conformation)
To Be Published
|
|
8JM8
 
 | | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with (R)-2-(2,2-dimethyl-4H-benzo[d][1,3]dioxin-6-yl)-2-hydroxyacetonitrile | | Descriptor: | (R)-2-(2,2-dimethyl-4H-benzo[d][1,3]dioxin-6-yl)-2-hydroxyacetonitrile, (R)-mandelonitrile lyase, 1,2-ETHANEDIOL, ... | | Authors: | Zheng, Y.-C, Li, F.-L, Yu, H.-L, Xu, J.-H. | | Deposit date: | 2023-06-04 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with (R)-2-(2,2-dimethyl-4H-benzo[d][1,3]dioxin-6-yl)-2-hydroxyacetonitrile
To Be Published
|
|
8JM4
 
 | | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 2-methyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde | | Descriptor: | (2S)-2-methyl-4H-1,3-benzodioxine-6-carbaldehyde, (R)-mandelonitrile lyase, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Y.-C, Li, F.-L, Yu, H.-L, Xu, J.-H. | | Deposit date: | 2023-06-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 2-methyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde
To Be Published
|
|
8JM7
 
 | | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A/S333V/P340L from Prunus communis complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde (noncatalytic conformation) | | Descriptor: | (R)-mandelonitrile lyase, 2,2-dimethyl-4H-1,3-benzodioxine-6-carbaldehyde, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Y.-C, Li, F.-L, Yu, H.-L, Xu, J.-H. | | Deposit date: | 2023-06-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant from Prunus communis mutant L331A complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde (Form A)
To Be Published
|
|
8JM3
 
 | | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 4H-benzo[d][1,3]dioxine-6-carbaldehyde | | Descriptor: | (R)-mandelonitrile lyase, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4H-1,3-benzodioxine-6-carbaldehyde, ... | | Authors: | Zheng, Y.-C, Li, F.-L, Yu, H.-L, Xu, J.-H. | | Deposit date: | 2023-06-04 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 4H-benzo[d][1,3]dioxine-6-carbaldehyde
To Be Published
|
|
8JM1
 
 | | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde from the cyanohydrin cleavage | | Descriptor: | (R)-mandelonitrile lyase, 1,2-ETHANEDIOL, 2,2-dimethyl-4H-1,3-benzodioxine-6-carbaldehyde, ... | | Authors: | Zheng, Y.-C, Li, F.-L, Yu, H.-L, Xu, J.-H. | | Deposit date: | 2023-06-04 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde from the cyanohydrin cleavage
To Be Published
|
|
8JM2
 
 | | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde (Form B) | | Descriptor: | (R)-mandelonitrile lyase, 1,2-ETHANEDIOL, 2,2-dimethyl-4H-1,3-benzodioxine-6-carbaldehyde, ... | | Authors: | Zheng, Y.-C, Li, F.-L, Yu, H.-L, Xu, J.-H. | | Deposit date: | 2023-06-04 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde (Form B)
To Be Published
|
|
8JM0
 
 | | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde (Form A) | | Descriptor: | (R)-mandelonitrile lyase, 2,2-dimethyl-4H-1,3-benzodioxine-6-carbaldehyde, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Y.-C, Li, F.-L, Yu, H.-L, Xu, J.-H. | | Deposit date: | 2023-06-04 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant from Prunus communis mutant L331A complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde (Form A)
To Be Published
|
|
