5IZP
 
 | | Solution Structure of DNA Dodecamer with 8-oxoguanine at 10th Position | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*(8OG)P*CP*G)-3') | | 著者 | Gruber, D.R, Hoppins, J.J, Miears, H.L, Kiryutin, A.S, Kasymov, R.D, Yurkovskaya, A.V, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2016-03-25 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 8-Oxoguanine Affects DNA Backbone Conformation in the EcoRI Recognition Site and Inhibits Its Cleavage by the Enzyme.
Plos One, 11, 2016
|
|
6FBU
 
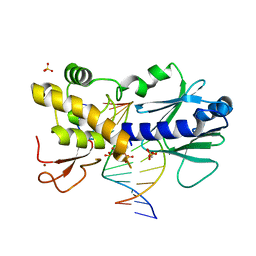 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli (E2Q) in complex with AP-site containing DNA substrate | | 分子名称: | ACETATE ION, DNA (5'-D(P*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3'), DNA (5'-D(P*GP*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*G)-3'), ... | | 著者 | Pomyalov, S, Lansky, S, Golan, G, Zharkov, D.O, Grollman, A.P, Shoham, G. | | 登録日 | 2017-12-19 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli (E2Q) in complex with AP-site containing DNA substrate
To Be Published
|
|
2FCC
 
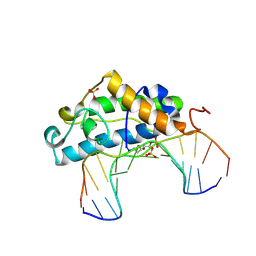 | | Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with a DNA Substrate Containing Abasic Site | | 分子名称: | DNA (5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*(BRU)P*(BRU)P*CP*AP*(BRU)P*CP*CP*(BRU)P*GP*G)-3'), Endonuclease V, ... | | 著者 | Golan, G, Zharkov, D.O, Fernandes, A.S, Dodson, M.L, McCullough, A.K, Grollman, A.P, Lloyd, R.S, Shoham, G. | | 登録日 | 2005-12-12 | | 公開日 | 2006-10-03 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of T4 Pyrimidine Dimer Glycosylase in a Reduced Imine Covalent Complex with Abasic Site-containing DNA.
J.Mol.Biol., 362, 2006
|
|
5IV1
 
 | | Solution Structure of DNA Dodecamer with 8-oxoguanine at 4th Position | | 分子名称: | DNA (5'-D(*CP*GP*CP*(8OG)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | 著者 | Miears, H.L, Gruber, D.R, Hoppins, J.J, Kiryutin, A.S, Kasymov, R.D, Yurkovskaya, A.V, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2016-03-18 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 8-Oxoguanine Affects DNA Backbone Conformation in the EcoRI Recognition Site and Inhibits Its Cleavage by the Enzyme.
Plos One, 11, 2016
|
|
2OQ4
 
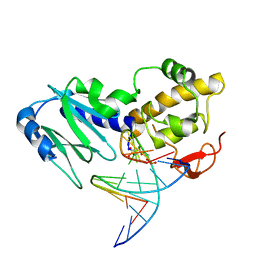 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli (E2Q) in complex with AP-site containing DNA substrate | | 分子名称: | 5'-D(*C*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*G*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*GP*G)-3', Endonuclease VIII, ... | | 著者 | Golan, G, Zharkov, D.O, Grollman, A.P, Shoahm, G. | | 登録日 | 2007-01-31 | | 公開日 | 2008-02-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Active site plasticity of endonuclease VIII: Conformational changes compensating for different substrates and mutations of critical residues
To be Published
|
|
5L06
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd Position | | 分子名称: | DNA (5'-D(*CP*GP*(5CM)P*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | 著者 | Miears, H.L, Hoppins, J.J, Gruber, D.R, Kasymov, R.D, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2016-07-26 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5L2G
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 9th Position | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(5CM)P*GP*CP*G)-3') | | 著者 | Miears, H.L, Hoppins, J.J, Gruber, D.R, Kasymov, R.D, Johnson, E.C, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2016-08-01 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
6ALT
 
 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 9th position | | 分子名称: | DNA (5'-D(*(DC5)P*GP*(DMC)P*GP*AP*AP*TP*TP*(DMC)P*GP*CP*(DG3))-3') | | 著者 | Gruber, D.R, Hoppins, J.J, Miears, H.L, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2017-08-08 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
6ALU
 
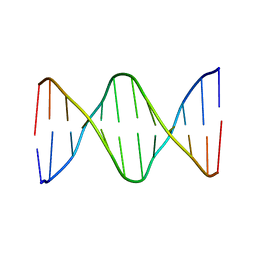 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 8-oxoguanine at the 4th position | | 分子名称: | DNA (5'-D(*(DC5)P*GP*(DMC)P*(8OG)P*AP*AP*TP*TP*CP*GP*CP*(DG3))-3') | | 著者 | Gruber, D.R, Shernyukov, A.V, Endutkin, A.V, Bagryanskaya, E.G, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2017-08-08 | | 公開日 | 2017-09-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
6ALS
 
 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 9th position and 8-oxoguanine at the 4th position | | 分子名称: | DNA (5'-D(*(DC5)P*GP*(DMC)P*(8OG)P*AP*AP*TP*TP*(DMC)P*GP*CP*(DG3))-3') | | 著者 | Gruber, D.R, Shernyukov, A.V, Endutkin, A.V, Bagryanskaya, E.G, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2017-08-08 | | 公開日 | 2017-09-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
1Q3B
 
 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli: The R252A mutant at 2.05 resolution. | | 分子名称: | Endonuclease VIII, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Golan, G, Zharkov, D.O, Feinberg, H, Fernandes, A.S, Zaika, E.I, Kycia, J.H, Grollman, A.P, Shoham, G. | | 登録日 | 2003-07-29 | | 公開日 | 2004-08-03 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility.
Nucleic Acids Res., 33, 2005
|
|
1Q39
 
 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli: The WT enzyme at 2.8 resolution. | | 分子名称: | CALCIUM ION, Endonuclease VIII, ZINC ION | | 著者 | Golan, G, Zharkov, D.O, Feinberg, H, Fernandes, A.S, Zaika, E.I, Kycia, J.H, Grollman, A.P, Shoham, G. | | 登録日 | 2003-07-29 | | 公開日 | 2004-08-03 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility.
Nucleic Acids Res., 33, 2005
|
|
1Q3C
 
 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli: The E2A mutant at 2.3 resolution. | | 分子名称: | Endonuclease VIII, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Golan, G, Zharkov, D.O, Feinberg, H, Fernandes, A.S, Zaika, E.I, Kycia, J.H, Grollman, A.P, Shoham, G. | | 登録日 | 2003-07-29 | | 公開日 | 2004-08-03 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility.
Nucleic Acids Res., 33, 2005
|
|
5TRN
 
 | | Solution Structure of a DNA Dodecamer with 8-oxoguanine at the 4th position and 5-methylcytosine at the 9th position | | 分子名称: | DNA (5'-D(*CP*GP*CP*(8OG)P*AP*AP*TP*TP*(DMC)P*GP*CP*G)-3') | | 著者 | Hoppins, J.J, Gruber, D.R, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2016-10-26 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5UZ3
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 9th position and 8-oxoguanine at the 10th position | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(DMC)P*(8OG)P*CP*G)-3') | | 著者 | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2017-02-24 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5UZ1
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd position and 8-oxoguanine at the 10th position | | 分子名称: | DNA (5'-D(*CP*GP*(DMC)P*GP*AP*AP*TP*TP*CP*(8OG)P*CP*G)-3') | | 著者 | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2017-02-24 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5UZ2
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd and 9th position and 8-oxoguanine at the 10th position | | 分子名称: | DNA (5'-D(*CP*GP*(DMC)P*GP*AP*AP*TP*TP*(DMC)P*(8OG)P*CP*G)-3') | | 著者 | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2017-02-24 | | 公開日 | 2017-03-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
1KG5
 
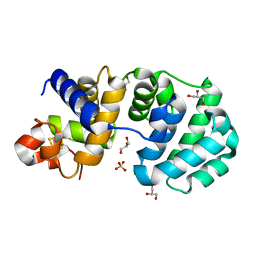 | | Crystal structure of the K142Q mutant of E.coli MutY (core fragment) | | 分子名称: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | 著者 | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | 登録日 | 2001-11-26 | | 公開日 | 2002-11-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG6
 
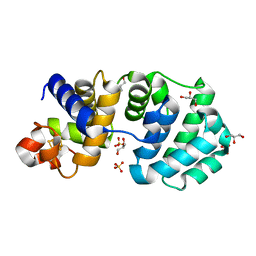 | | Crystal structure of the K142R mutant of E.coli MutY (core fragment) | | 分子名称: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | 著者 | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | 登録日 | 2001-11-26 | | 公開日 | 2002-11-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG4
 
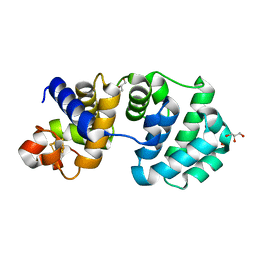 | | Crystal structure of the K142A mutant of E. coli MutY (core fragment) | | 分子名称: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | 著者 | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | 登録日 | 2001-11-26 | | 公開日 | 2002-11-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG2
 
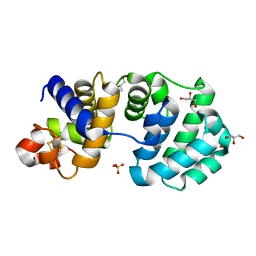 | | Crystal structure of the core fragment of MutY from E.coli at 1.2A resolution | | 分子名称: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | 著者 | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | 登録日 | 2001-11-26 | | 公開日 | 2002-11-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG7
 
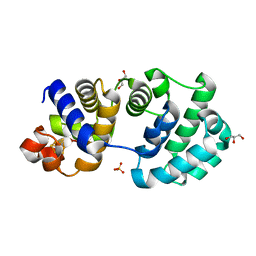 | | Crystal Structure of the E161A mutant of E.coli MutY (core fragment) | | 分子名称: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | 著者 | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | 登録日 | 2001-11-26 | | 公開日 | 2002-11-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG3
 
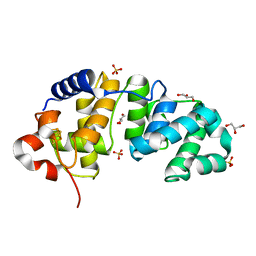 | | Crystal structure of the core fragment of MutY from E.coli at 1.55A resolution | | 分子名称: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | 著者 | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | 登録日 | 2001-11-26 | | 公開日 | 2002-11-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1K3W
 
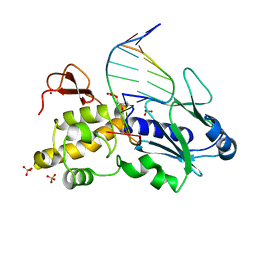 | | Crystal structure of a trapped reaction intermediate of the DNA Repair Enzyme Endonuclease VIII with DNA | | 分子名称: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*GP*G)-3', Endonuclease VIII, ... | | 著者 | Golan, G, Zharkov, D.O, Gilboa, R, Fernandes, A.S, Kycia, J.H, Gerchman, S.E, Rieger, R.A, Grollman, A.P, Shoham, G. | | 登録日 | 2001-10-04 | | 公開日 | 2002-10-04 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate.
EMBO J., 21, 2002
|
|
1K3X
 
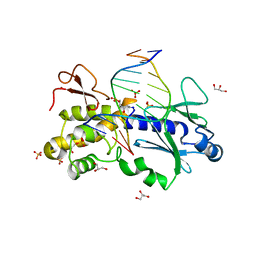 | | Crystal structure of a trapped reaction intermediate of the DNA repair enzyme Endonuclease VIII with Brominated-DNA | | 分子名称: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*(BRU)P*(BRU)P*CP*AP*(BRU)P*CP*CP*(BRU)P*GP*G)-3', Endonuclease VIII, ... | | 著者 | Golan, G, Zharkov, D.O, Gilboa, R, Fernandes, A.S, Kycia, J.H, Gerchman, S.E, Rieger, R.A, Grollman, A.P, Shoham, G. | | 登録日 | 2001-10-04 | | 公開日 | 2002-10-04 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate.
EMBO J., 21, 2002
|
|
