4JBK
 
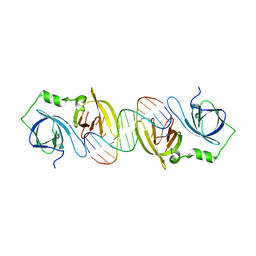 | | Molecular basis for abrogation of activation of pro-inflammatory cytokines | | Descriptor: | DNA (5'-D(P*GP*GP*AP*AP*TP*TP*AP*TP*AP*AP*TP*TP*CP*C)-3'), Interferon-activable protein 202 | | Authors: | Ru, H, Ni, X, Crowley, C, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
2KCQ
 
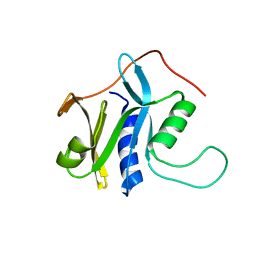 | | Solution structure of protein SRU_2040 from Salinibacter ruber (strain DSM 13855) . Northeast Structural Genomics Consortium target SrR106 | | Descriptor: | Mov34/MPN/PAD-1 family | | Authors: | Wu, Y, Eletsky, A, Zhao, L, Hua, J, Sukumaran, D, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-28 | | Release date: | 2009-02-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein SRU_2040 from Salinibacter ruber (strain DSM 13855). Northeast Structural Genomics Consortium target SrR106
To be Published
|
|
8XR6
 
 | | Cryo-EM structure of cryptophyte photosystem II | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, ... | | Authors: | Li, K, Zhao, L.S, Zhang, Y.Z, Liu, L.N. | | Deposit date: | 2024-01-06 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Cryo-EM structure of cryptophyte photosystem II
To Be Published
|
|
4NZG
 
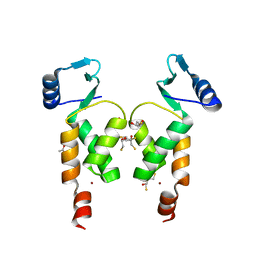 | | Crystal Structure of the N-terminal domain of Moloney murine leukemia virus integrase, Northeast Structural Genomics Consortium Target OR3 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Integrase p46, ... | | Authors: | Guan, R, Jiang, M, Janjua, H, Maglaqui, M, Zhao, L, Xiao, R, Acton, T.B, Everett, J.K, Roth, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-12-12 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | X-ray crystal structure of the N-terminal region of Moloney murine leukemia virus integrase and its implications for viral DNA recognition.
Proteins, 85, 2017
|
|
4KWE
 
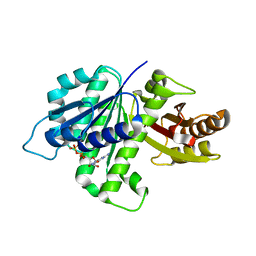 | | GDP-bound, double-stranded, curved FtsZ protofilament structure | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Li, Y, Hsin, J, Zhao, L, Cheng, Y, Shang, W, Huang, K.C, Wang, H.W, Ye, S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation
Science, 341, 2013
|
|
7F16
 
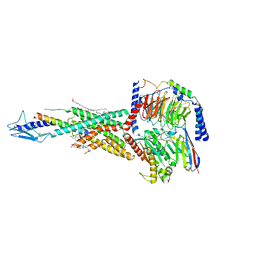 | | Cryo-EM structure of parathyroid hormone receptor type 2 in complex with a tuberoinfundibular peptide of 39 residues and G protein | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, X, Cheng, X, Zhao, L.H, Wang, Y, Ye, C, Zou, X, Dai, A, Cong, Z, Chen, J, Zhou, Q, Xia, T, Jiang, H, Xu, H.E, Yang, D, Wang, M.W. | | Deposit date: | 2021-06-08 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular insights into differentiated ligand recognition of the human parathyroid hormone receptor 2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7V9M
 
 | | Cryo-EM structure of the GHRH-bound human GHRHR splice variant 1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Chen, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7V9L
 
 | | Cryo-EM structure of the SV1-Gs complex. | | Descriptor: | GHRH receptor splice variant 1,GHRH receptor splice variant 1,GHRH receptor splice variant 1,SV1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Cheng, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2022-04-06 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DUQ
 
 | | Cryo-EM structure of the compound 2 and GLP-1-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7DUR
 
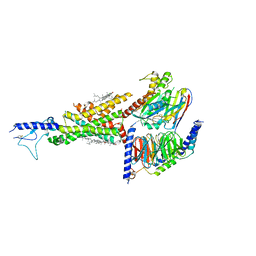 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7WOT
 
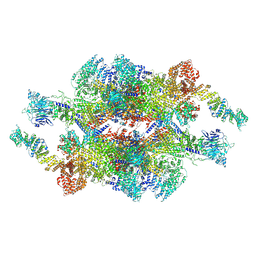 | | Cryo-EM structure of the inner ring monomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7WOO
 
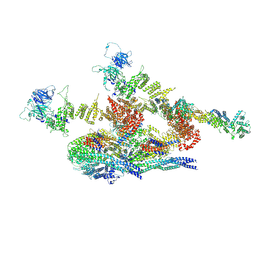 | | Cryo-EM structure of the inner ring protomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7EVM
 
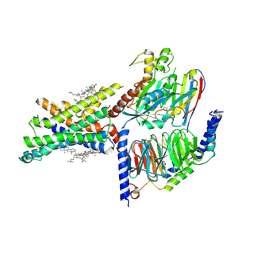 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-05-21 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7DKA
 
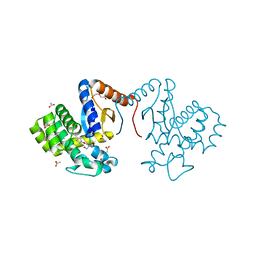 | | Crystal structure of DsbA-like protein DR2335 from Deinococcus radiodurans R1, C24S mutant protein | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, M.-K, Zhang, J, Zhao, L. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of DsbA-like protein DR2335 from Deinococcus radiodurans R1, native protein
To Be Published
|
|
7WQR
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 28 | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ~{N}-(3-chlorophenyl)-2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]benzamide | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7WQM
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 24 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(2-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7WQS
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 25 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(3-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7XH0
 
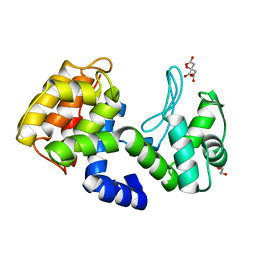 | | crystal structure of Csn-PD from Paenibacillus dendritiformis | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Chitosanase | | Authors: | Sun, H.H, Cheng, Y.M, Cao, R, Liu, Q, Zhao, L. | | Deposit date: | 2022-04-07 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | crystal structure of Csn-PD from Paenibacillus dendritiformis
To Be Published
|
|
2HD3
 
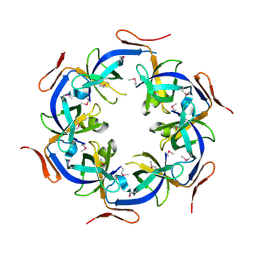 | | Crystal Structure of the Ethanolamine Utilization Protein EutN from Escherichia coli, NESG Target ER316 | | Descriptor: | Ethanolamine utilization protein eutN | | Authors: | Forouhar, F, Zhang, W, Jayaraman, S, Zhao, L, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-19 | | Release date: | 2006-08-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2HQV
 
 | | X-ray Crystal Structure of Protein AGR_C_4470 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR92. | | Descriptor: | AGR_C_4470p | | Authors: | Vorobiev, S.M, Neely, H, Seetharaman, J, Zhao, L, Cunningham, K, Ma, L.C, Fang, Y, Xiao, R, Acton, T, Montelione, T.G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-19 | | Release date: | 2006-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of AGR_C_4470p from Agrobacterium tumefaciens.
Protein Sci., 16, 2007
|
|
7DK9
 
 | |
7D6L
 
 | |
2ICT
 
 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 8.5. Northeast Structural Genomics TARGET ER390. | | Descriptor: | antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
2JZ5
 
 | | NMR solution structure of protein VPA0419 from Vibrio parahaemolyticus. Northeast Structural Genomics target VpR68 | | Descriptor: | Uncharacterized protein VPA0419 | | Authors: | Singarapu, K.K, Sukumaran, D.K, Eletski, A, Parish, D, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of proteins VPA0419 from Vibrio parahaemolyticus and yiiS from Shigella flexneri provide structural coverage for protein domain family PFAM 04175.
Proteins, 78, 2010
|
|
2JRX
 
 | | Solution NMR structure of protein YejL from E. coli. Northeast Structural Genomics target ER309 | | Descriptor: | UPF0352 protein yejL | | Authors: | Cort, J.R, Zhao, L, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Chemical Shift Assignments of E. coli YejL protein.
To be Published
|
|
