8GTP
 
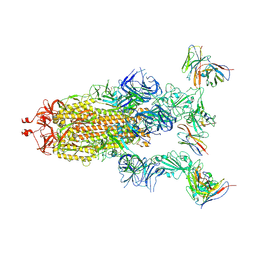 | | cryo-EM structure of Omicron BA.5 S protein in complex with XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
8GTQ
 
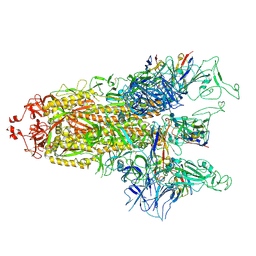 | | cryo-EM structure of Omicron BA.5 S protein in complex with S2L20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
7TVK
 
 | |
8HNC
 
 | | Cryo-EM structure of human OATP1B1 in complex with bilirubin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Shan, Z, Yang, X, Liu, H, Nan, J, Yuan, Y, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
8HNB
 
 | | Cryo-EM structure of human OATP1B1 in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier organic anion transporter family member 1B1 | | Authors: | Shan, Z, Yang, X, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
8HNH
 
 | | Cryo-EM structure of human OATP1B1 in complex with simeprevir | | Descriptor: | (2R,3aR,10Z,11aS,12aR,14aR)-N-(cyclopropylsulfonyl)-2-({7-methoxy-8-methyl-2-[4-(1-methylethyl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-4,14-dioxo-2,3,3a,4,5,6,7,8,9,11a,12,13,14,14a-tetradecahydrocyclopenta[c]cyclopropa[g][1,6]diazacyclotetradecine-12a(1H)-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shan, Z, Yang, X, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
8HND
 
 | | Cryo-EM structure of human OATP1B1 in complex with estrone-3-sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier organic anion transporter family member 1B1, ... | | Authors: | Shan, Z, Yang, X, Liu, H, Nan, J, Yuan, Y, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
5ZB5
 
 | | The structural basis of histone chaperoneVps75 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NAP family histone chaperone vps75 | | Authors: | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | Deposit date: | 2018-02-09 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
5ZIT
 
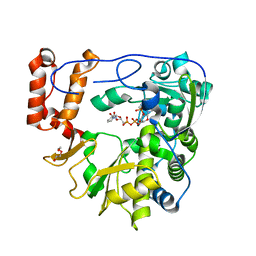 | | Crystal structure of human Enterovirus D68 RdRp in complex with NADPH | | Descriptor: | DI(HYDROXYETHYL)ETHER, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RdRp | | Authors: | Wang, M.L, Li, L, Chen, Y.P, Jiang, H, Zhang, Y, Su, D. | | Deposit date: | 2018-03-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 2020
|
|
7CT5
 
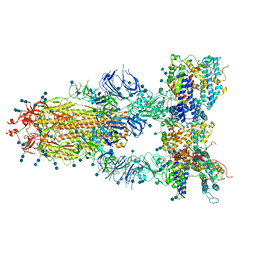 | | S protein of SARS-CoV-2 in complex bound with T-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Guo, L, Bi, W.W, Zhang, Y.Y, Yan, R.H, Li, Y.N, Zhou, Q, Dang, B.B. | | Deposit date: | 2020-08-18 | | Release date: | 2020-11-18 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Engineered trimeric ACE2 binds viral spike protein and locks it in "Three-up" conformation to potently inhibit SARS-CoV-2 infection.
Cell Res., 31, 2021
|
|
8QQK
 
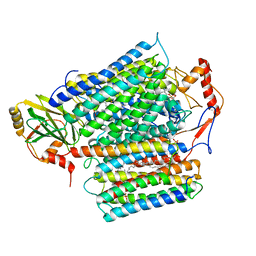 | | Cryo-EM structure of E. coli cytochrome bo3 quinol oxidase assembled in peptidiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, ... | | Authors: | Gao, Y, Zhang, Y, Hakke, S, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2023-10-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of cytochrome bo 3 quinol oxidase assembled in peptidiscs reveals an "open" conformation for potential ubiquinone-8 release.
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
5W96
 
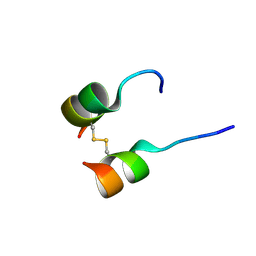 | | Solution structure of phage derived peptide inhibitor of frizzled 7 receptor | | Descriptor: | Fz7 binding peptide | | Authors: | Nile, A.H, de Sousa e Melo, F, Mukund, S, Piskol, R, Hansen, S, Zhou, L, Zhang, Y, Fu, Y, Gogol, E.B, Komuves, L.G, Modrusan, Z, Angers, S, Franke, Y, Koth, C, Fairbrother, W.J, Wang, W, de Sauvage, F.J, Hannoush, R.N. | | Deposit date: | 2017-06-22 | | Release date: | 2018-04-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
6UTS
 
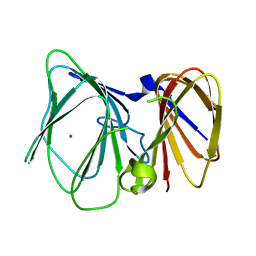 | |
8J86
 
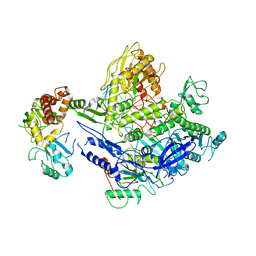 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*CP*TP*TP*AP*AP*TP*CP*TP*CP*AP*CP*AP*TP*AP*GP*CP*AP*GP*CP*TP*)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-04-30 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
8J8F
 
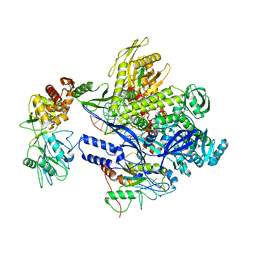 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 in complex with a DNA duplex and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-05-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
8J8G
 
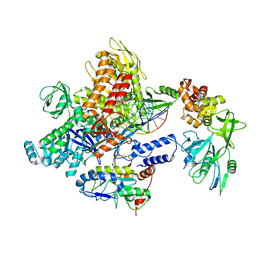 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 in complex with a DNA duplex and cidofovir diphosphate | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*CP*TP*TP*AP*AP*TP*CP*TP*CP*AP*CP*AP*TP*AP*GP*CP*AP*GP*CP*T)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-05-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
8K14
 
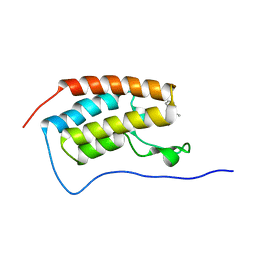 | | X-ray crystal structure of 18a in BRD4(1) | | Descriptor: | 4-[8-methoxy-2-methyl-1-(1-phenylethyl)imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 4 | | Authors: | Xu, H, Shen, H, Zhang, Y, Xu, Y, Li, R. | | Deposit date: | 2023-07-10 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Discovery of (R)-4-(8-methoxy-2-methyl-1-(1-phenylethy)-1H-imidazo[4,5-c]quinnolin-7-yl)-3,5-dimethylisoxazole as a potent and selective BET inhibitor for treatment of acute myeloid leukemia (AML) guided by FEP calculation.
Eur.J.Med.Chem., 263, 2024
|
|
8J6R
 
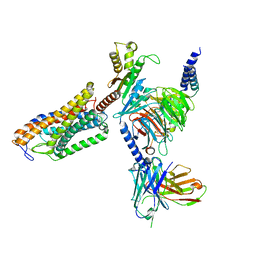 | | Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
8J6Q
 
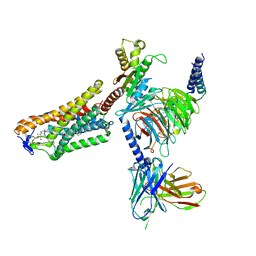 | | Cryo-EM structure of the 3-HB and compound 9n-bound human HCAR2-Gi1 complex | | Descriptor: | (3R)-3-hydroxybutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
8J6P
 
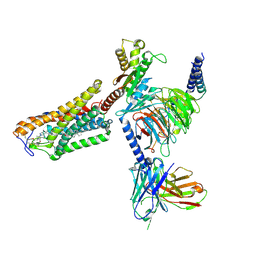 | | Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
6URG
 
 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-CPSF100 PIM complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 4, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
5XDZ
 
 | | Crystal structure of zebrafish SNX25 PX domain | | Descriptor: | CHLORIDE ION, Cellular trafficking protein, SODIUM ION | | Authors: | Su, K, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the PX domain of SNX25 reveals a novel phospholipid recognition model by dimerization in the PX domain
FEBS Lett., 591, 2017
|
|
6URO
 
 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-PAS RNA-CstF77 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, Cleavage stimulation factor subunit 3, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
5YIU
 
 | | Caulobacter crescentus GcrA DNA-binding domain (DBD) | | Descriptor: | Cell cycle regulatory protein GcrA | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
5YIW
 
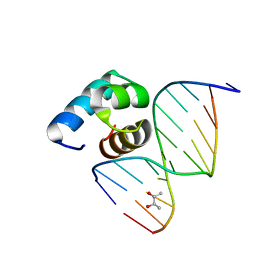 | | Caulobacter crescentus GcrA DNA-binding domain (DBD) in complex with methylated dsDNA (crystal form 2) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cell cycle regulatory protein GcrA, DNA (5'-D(*CP*CP*CP*TP*GP*(6MA)P*TP*TP*CP*GP*C)-3'), ... | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
