5WMG
 
 | | N-terminal bromodomain of BRD4 in complex with OTX-015 | | 分子名称: | 1,2-ETHANEDIOL, 4-{6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1S)-1-(pyridin-2-yl)ethyl]-1H-pyrrolo[3,2-b]pyridin-3-yl}benzoic acid, Bromodomain-containing protein 4 | | 著者 | Zhang, Y. | | 登録日 | 2017-07-28 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
5WMD
 
 | | N-terminal bromodomain of BRD4 in complex with OTX-015 | | 分子名称: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(4-hydroxyphenyl)acetamide, Bromodomain-containing protein 4 | | 著者 | Zhang, Y. | | 登録日 | 2017-07-28 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
5WMA
 
 | | N-terminal bromodomain of BRD4 in complex with PLX5981 | | 分子名称: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1H-pyrrolo[2,3-b]pyridine, Bromodomain-containing protein 4 | | 著者 | Zhang, Y. | | 登録日 | 2017-07-28 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.401 Å) | | 主引用文献 | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
7XRJ
 
 | | crystal structure of N-acetyltransferase DgcN-25328 | | 分子名称: | Putative NAD-dependent epimerase/dehydratase family protein, SULFATE ION | | 著者 | Zhang, Y.Z, Yu, Y, Cao, H.Y, Chen, X.L, Wang, P. | | 登録日 | 2022-05-10 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Novel D-glutamate catabolic pathway in marine Proteobacteria and halophilic archaea.
Isme J, 17, 2023
|
|
6KZK
 
 | |
6C00
 
 | |
4OXW
 
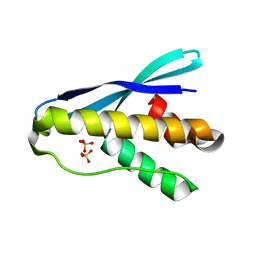 | |
2MNZ
 
 | | NMR Structure of KDM5B PHD1 finger in complex with H3K4me0(1-10aa) | | 分子名称: | H3K4me0, Lysine-specific demethylase 5B, ZINC ION | | 著者 | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | 登録日 | 2014-04-16 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
2MK6
 
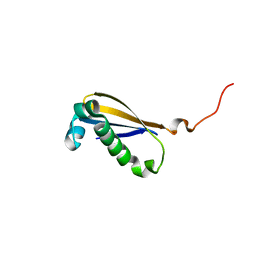 | |
6MG8
 
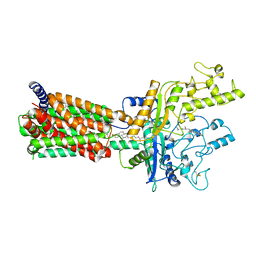 | | Structural basis for cholesterol transport-like activity of the Hedgehog receptor Patched | | 分子名称: | CHOLESTEROL, Protein patched homolog 1 | | 著者 | Zhang, Y, Bulkley, D, Xin, Y, Roberts, K.J, Asarnow, D.E, Sharma, A, Myers, B.R, Cho, W, Cheng, Y, Beachy, P.A. | | 登録日 | 2018-09-13 | | 公開日 | 2018-11-28 | | 最終更新日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched.
Cell, 175, 2018
|
|
6LPI
 
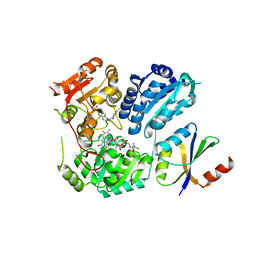 | | Crystal Structure of AHAS holo-enzyme | | 分子名称: | Acetolactate synthase isozyme 1 large subunit, Acetolactate synthase isozyme 1 small subunit, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Zhang, Y, Yang, X, Xi, Z, Shen, Y. | | 登録日 | 2020-01-10 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.849 Å) | | 主引用文献 | Molecular architecture of the acetohydroxyacid synthase holoenzyme.
Biochem.J., 477, 2020
|
|
2MNY
 
 | | NMR Structure of KDM5B PHD1 finger | | 分子名称: | Lysine-specific demethylase 5B, ZINC ION | | 著者 | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | 登録日 | 2014-04-16 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
8UAI
 
 | |
1Z34
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine | | 分子名称: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z33
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase | | 分子名称: | purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z39
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2'-deoxyinosine | | 分子名称: | 2'-DEOXYINOSINE, purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z38
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with inosine | | 分子名称: | INOSINE, purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z35
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoroadenosine | | 分子名称: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
5HMC
 
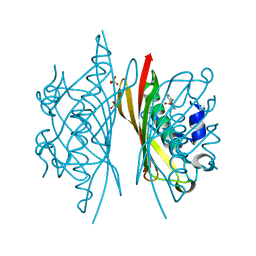 | | Crystal structure of S. sahachiroi AziG complexed with 5-methyl naphthoic acid | | 分子名称: | 5-methylnaphthalene-1-carboxylic acid, Azi13, SULFATE ION | | 著者 | Zhang, Y, Erb, M.S, Ealick, S.E. | | 登録日 | 2016-01-15 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Polyketide Ring Expansion Mediated by a Thioesterase, Chain Elongation and Cyclization Domain, in Azinomycin Biosynthesis: Characterization of AziB and AziG.
Biochemistry, 55, 2016
|
|
1Z37
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with adenosine | | 分子名称: | ADENOSINE, purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z36
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with formycin A | | 分子名称: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
6LTS
 
 | |
8CZJ
 
 | | A bacteria Zrt/Irt-like protein in the apo state | | 分子名称: | Putative membrane protein, SULFATE ION, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | 著者 | Zhang, Y, Hu, J. | | 登録日 | 2022-05-24 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.751 Å) | | 主引用文献 | Structural insights into the elevator-type transport mechanism of a bacterial ZIP metal transporter.
Nat Commun, 14, 2023
|
|
6KQD
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-OH RNA of 3 nt | | 分子名称: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), ... | | 著者 | Zhang, Y, Li, L, Ebright, R.H. | | 登録日 | 2019-08-17 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y5B
 
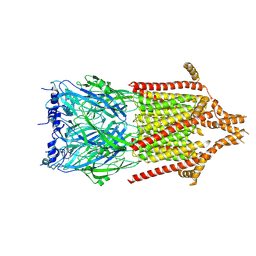 | | 5-HT3A receptor in Salipro (apo, asymmetric) | | 分子名称: | 5-hydroxytryptamine receptor 3A | | 著者 | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | 登録日 | 2020-02-25 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
