5LNR
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-PLP complex | | 分子名称: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal 5'-phosphate synthase subunit PDX1.3 | | 著者 | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | 登録日 | 2016-08-06 | | 公開日 | 2017-01-18 | | 最終更新日 | 2017-02-22 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LNT
 
 | | Crystal structure of Arabidopsis thaliana Pdx1K166R-preI320 complex | | 分子名称: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.1, [(~{E},4~{S})-4-azanyl-3-oxidanylidene-pent-1-enyl] dihydrogen phosphate | | 著者 | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | 登録日 | 2016-08-06 | | 公開日 | 2017-01-18 | | 最終更新日 | 2017-02-22 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
1SPX
 
 | | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form | | 分子名称: | short-chain reductase family member (5L265) | | 著者 | Schormann, N, Zhou, J, McCombs, D, Bray, T, Symersky, J, Huang, W.-Y, Luan, C.-H, Gray, R, Luo, D, Arabashi, A, Bunzel, B, Nagy, L, Lu, S, Li, S, Lin, G, Zhang, Y, Qiu, S, Tsao, J, Luo, M, Carson, M, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2004-03-17 | | 公開日 | 2004-03-23 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form: A Member of the SDR-Family
To be Published
|
|
1P51
 
 | | Anabaena HU-DNA cocrystal structure (AHU6) | | 分子名称: | 5'-D(*GP*CP*AP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*T)-3', DNA-binding protein HU | | 著者 | Swinger, K.K, Lemberg, K.M, Zhang, Y, Rice, P.A. | | 登録日 | 2003-04-24 | | 公開日 | 2003-05-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Flexible DNA bending in HU-DNA cocrystal structures
Embo J., 22, 2003
|
|
1PFB
 
 | | Structural Basis for specific binding of polycomb chromodomain to histone H3 methylated at K27 | | 分子名称: | ACETIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Min, J.R, Zhang, Y, Xu, R.-M. | | 登録日 | 2003-05-24 | | 公開日 | 2003-10-07 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27.
Genes Dev., 17, 2003
|
|
1T7S
 
 | | Structural Genomics of Caenorhabditis elegans: Structure of BAG-1 protein | | 分子名称: | BAG-1 cochaperone | | 著者 | Symersky, J, Zhang, Y, Schormann, N, Li, S, Bunzel, R, Pruett, P, Luan, C.-H, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2004-05-10 | | 公開日 | 2004-05-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural genomics of Caenorhabditis elegans: structure of the BAG domain.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SZP
 
 | | A Crystal Structure of the Rad51 Filament | | 分子名称: | DNA repair protein RAD51, SULFATE ION | | 著者 | Conway, A.B, Lynch, T.W, Zhang, Y, Fortin, G.S, Symington, L.S, Rice, P.A. | | 登録日 | 2004-04-06 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Crystal structure of a Rad51 filament.
Nat.Struct.Mol.Biol., 11, 2004
|
|
7F0R
 
 | | Cryo-EM structure of Pseudomonas aeruginosa SutA transcription activation complex | | 分子名称: | DNA (70-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | He, D.W, You, L.L, Zhang, Y. | | 登録日 | 2021-06-06 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
6WXK
 
 | | PHF23 PHD Domain Apo | | 分子名称: | PHD finger protein 23, ZINC ION | | 著者 | Vann, K.R, Zhang, J, Zhang, Y, Kutateladze, T. | | 登録日 | 2020-05-11 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Mechanistic insights into chromatin targeting by leukemic NUP98-PHF23 fusion.
Nat Commun, 11, 2020
|
|
7JVR
 
 | | Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in complex with Gi protein | | 分子名称: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | 登録日 | 2020-08-22 | | 公開日 | 2021-02-24 | | 最終更新日 | 2021-03-31 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
3JA8
 
 | | Cryo-EM structure of the MCM2-7 double hexamer | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Minichromosome Maintenance 2, Minichromosome Maintenance 3, ... | | 著者 | Li, N, Zhai, Y, Zhang, Y, Li, W, Yang, M, Lei, J, Tye, B.K, Gao, N. | | 登録日 | 2015-05-09 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the eukaryotic MCM complex at 3.8 angstrom
Nature, 524, 2015
|
|
7JVQ
 
 | | Cryo-EM structure of apomorphine-bound dopamine receptor 1 in complex with Gs protein | | 分子名称: | (6aR)-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | 著者 | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | 登録日 | 2020-08-22 | | 公開日 | 2021-02-24 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVP
 
 | | Cryo-EM structure of SKF-83959-bound dopamine receptor 1 in complex with Gs protein | | 分子名称: | (1R)-6-chloro-3-methyl-1-(3-methylphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | 著者 | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | 登録日 | 2020-08-22 | | 公開日 | 2021-02-24 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JV5
 
 | | Cryo-EM structure of SKF-81297-bound dopamine receptor 1 in complex with Gs protein | | 分子名称: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | 著者 | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | 登録日 | 2020-08-20 | | 公開日 | 2021-02-24 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
3KVM
 
 | |
3L5T
 
 | |
5Y8Y
 
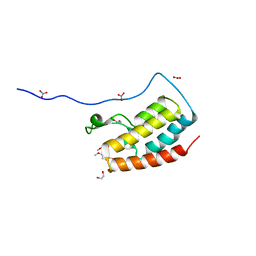 | | Crystal Structure Analysis of the BRD4 | | 分子名称: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | 著者 | Xu, Y, Zhang, Y, Song, M, Wang, C. | | 登録日 | 2017-08-21 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
8JCH
 
 | |
3L5R
 
 | |
3L5U
 
 | |
3L5S
 
 | |
3L5V
 
 | |
3L5P
 
 | |
6UTS
 
 | |
3JSG
 
 | |
