1VAZ
 
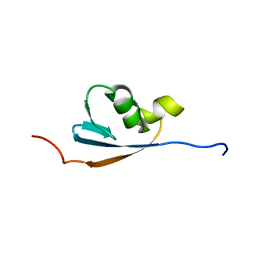 | | Solution structures of the p47 SEP domain | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-02-20 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97.
Embo J., 23, 2004
|
|
5EZK
 
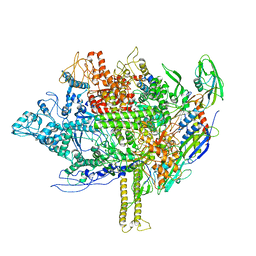 | | RNA polymerase model placed by Molecular replacement into X-ray diffraction map of DNA-bound RNA Polymerase-Sigma 54 holoenzyme complex. | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darbari, V.C, Yang, Y, Lu, D, Zhang, N, Glyde, R, Wang, Y, Murakami, K.S, Buck, M, Zhang, X. | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.5 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase- Sigma 54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
1V92
 
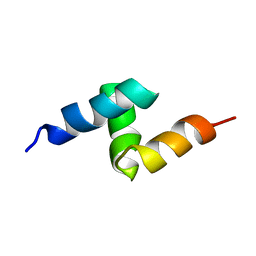 | | Solution structure of the UBA domain from p47, a major cofactor of the AAA ATPase p97 | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-01-19 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97
Embo J., 23, 2004
|
|
1WE1
 
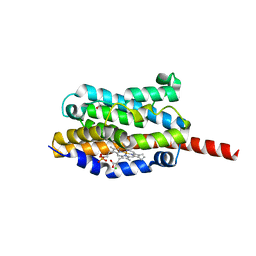 | | Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC6803 in complex with heme | | Descriptor: | CHLORIDE ION, Heme oxygenase 1, ISOPROPYL ALCOHOL, ... | | Authors: | Sugishima, M, Migita, C.T, Zhang, X, Yoshida, T, Fukuyama, K. | | Deposit date: | 2004-05-21 | | Release date: | 2004-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC 6803 in complex with heme
Eur.J.Biochem., 271, 2004
|
|
4TOR
 
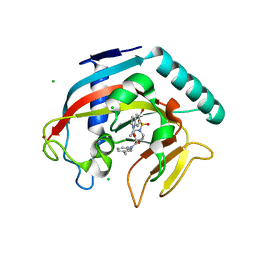 | | Crystal structure of Tankyrase 1 with IWR-8 | | Descriptor: | 1-[(1-acetyl-5-bromo-1H-indol-6-yl)sulfonyl]-N-ethyl-N-(3-methylphenyl)piperidine-4-carboxamide, CHLORIDE ION, Tankyrase-1, ... | | Authors: | Chen, H, Zhang, X, Lum, L, Chen, C. | | Deposit date: | 2014-06-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
4TOS
 
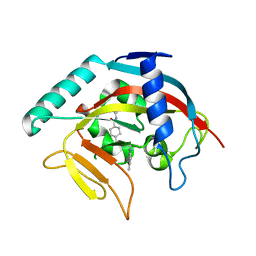 | | Crystal structure of Tankyrase 1 with 355 | | Descriptor: | Tankyrase-1, ZINC ION, trans-N-benzyl-4-({1-[(6-methyl-4-oxo-4H-pyrido[1,2-a]pyrimidin-2-yl)methyl]-2,4-dioxo-1,4-dihydroquinazolin-3(2H)-yl}methyl)cyclohexanecarboxamide | | Authors: | Chen, H, Zhang, X, Lum, l, Chen, C. | | Deposit date: | 2014-06-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
3JBL
 
 | | Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated Polymerization | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Zhang, L, Chen, S, Ruan, J, Wu, J, Tong, A.B, Yin, Q, Li, Y, David, L, Lu, A, Wang, W.L, Marks, C, Ouyang, Q, Zhang, X, Mao, Y, Wu, H. | | Deposit date: | 2015-09-05 | | Release date: | 2015-10-21 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization.
Science, 350, 2015
|
|
1EUE
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oganesyan, V, Zhang, X. | | Deposit date: | 2000-04-19 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of redox potential in electron transfer proteins: effects of complex formation on the active site microenvironment of cytochrome b5.
FARADAY DISC.CHEM.SOC, 116, 2001
|
|
1F9W
 
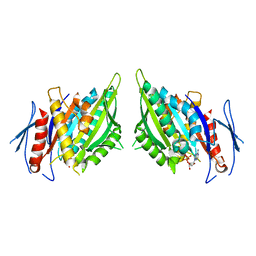 | | CRYSTAL STRUCTURES OF MUTANTS REVEAL A SIGNALLING PATHWAY FOR ACTIVATION OF THE KINESIN MOTOR ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | Authors: | Yun, M, Zhang, X, Park, C.G, Park, H.W, Endow, S.A. | | Deposit date: | 2000-07-11 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural pathway for activation of the kinesin motor ATPase.
EMBO J., 20, 2001
|
|
1F9T
 
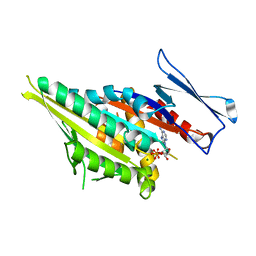 | | CRYSTAL STRUCTURES OF KINESIN MUTANTS REVEAL A SIGNALLING PATHWAY FOR ACTIVATION OF THE MOTOR ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | Authors: | Yun, M, Zhang, X, Park, C.-G, Park, H.-W, Endow, S.A. | | Deposit date: | 2000-07-11 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural pathway for activation of the kinesin motor ATPase.
EMBO J., 20, 2001
|
|
1F9V
 
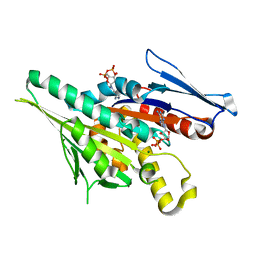 | | CRYSTAL STRUCTURES OF MUTANTS REVEAL A SIGNALLING PATHWAY FOR ACTIVATION OF THE KINESIN MOTOR ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | Authors: | Yun, M, Zhang, X, Park, C.G, Park, H.W, Endow, S.A. | | Deposit date: | 2000-07-11 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A structural pathway for activation of the kinesin motor ATPase.
EMBO J., 20, 2001
|
|
1FLC
 
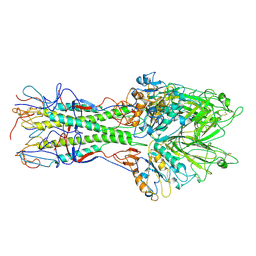 | | X-RAY STRUCTURE OF THE HAEMAGGLUTININ-ESTERASE-FUSION GLYCOPROTEIN OF INFLUENZA C VIRUS | | Descriptor: | HAEMAGGLUTININ-ESTERASE-FUSION GLYCOPROTEIN, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rosenthal, P.B, Zhang, X, Formanowski, F, Fitz, W, Wong, C.H, Meier-Ewert, H, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1999-02-22 | | Release date: | 2000-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus.
Nature, 396, 1998
|
|
1F9U
 
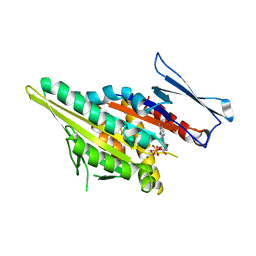 | | CRYSTAL STRUCTURES OF MUTANTS REVEAL A SIGNALLING PATHWAY FOR ACTIVATION OF THE KINESIN MOTOR ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | Authors: | Yun, M, Zhang, X, Park, C.G, Park, H.W, Endow, S.A. | | Deposit date: | 2000-07-11 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural pathway for activation of the kinesin motor ATPase.
EMBO J., 20, 2001
|
|
1FZX
 
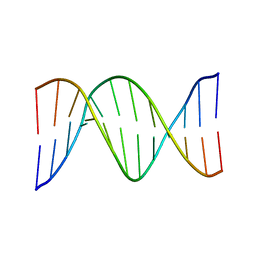 | | NMR SOLUTION STRUCTURE OF THE DNA DODECAMER GGCAAAAAACGG | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*TP*TP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*AP*AP*AP*AP*CP*GP*G)-3' | | Authors: | MacDonald, D, Herbert, K, Zhang, X, Pologruto, T, Lu, P. | | Deposit date: | 2000-10-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an A-tract DNA bend.
J.Mol.Biol., 306, 2001
|
|
1G14
 
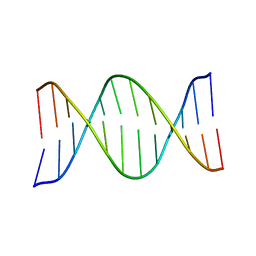 | | NMR SOLUTION STRUCTURE OF THE DNA DODECAMER GGCAAGAAACGG | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*TP*CP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*GP*AP*AP*AP*CP*GP*G)-3' | | Authors: | MacDonald, D, Herbert, K, Zhang, X, Pologruto, T, Lu, P. | | Deposit date: | 2000-10-10 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an A-tract DNA bend.
J.Mol.Biol., 306, 2001
|
|
5CP7
 
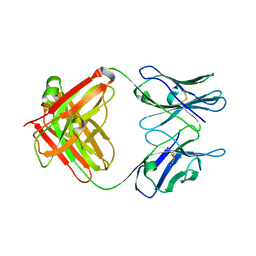 | | Crystal Structure of an Antigen-Binding Fragment of Monoclonal Antibody against Sulfonamides | | Descriptor: | Heavy Chain of Antigen-Binding Fragment of Monoclonal Antibody of 4C7, Light Chain of Antigen-Binding Fragment of Monoclonal Antibody of 4C7 | | Authors: | Wang, Z, Shen, J, Li, C, Li, Y, Wen, K, Yu, X, Zhang, X. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Class-specific Monoclonal Antibodies and Dihydropteroate Synthase in Bioassays used for the Detection of Sulfonamides: Structural Insights into Recognition Diversity.
Anal. Chem., 91, 2019
|
|
4V40
 
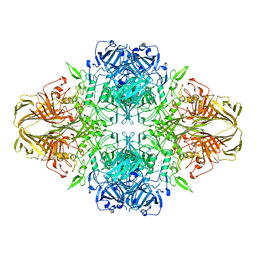 | | BETA-GALACTOSIDASE | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Jacobson, R.H, Zhang, X, Dubose, R.F, Matthews, B.W. | | Deposit date: | 1994-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of beta-galactosidase from E. coli.
Nature, 369, 1994
|
|
5CP3
 
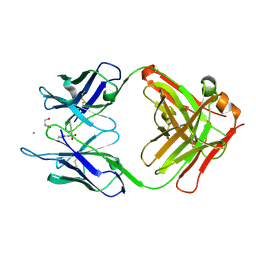 | | Crystal Structure of an Antigen-Binding Fragment of Monoclonal Antibody against Sulfonamides in Complex with Sulfathiazole | | Descriptor: | 4-amino-N-(1,3-thiazol-2-yl)benzenesulfonamide, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Z, Shen, J, Li, C, Li, Y, Wen, K, Yu, X, Zhang, X. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Class-specific Monoclonal Antibodies and Dihydropteroate Synthase in Bioassays used for the Detection of Sulfonamides: Structural Insights into Recognition Diversity.
Anal. Chem., 91, 2019
|
|
4ZDS
 
 | | Crystal Structure of core DNA binding domain of Arabidopsis Thaliana Transcription Factor Ethylene-Insensitive 3 | | Descriptor: | Protein ETHYLENE INSENSITIVE 3 | | Authors: | Song, J, Zhu, C, Zhang, X, Wen, X, Liu, L, Peng, J, Guo, H, Yi, C. | | Deposit date: | 2015-04-18 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Biochemical and Structural Insights into the Mechanism of DNA Recognition by Arabidopsis ETHYLENE INSENSITIVE3.
Plos One, 10, 2015
|
|
1P9E
 
 | | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3 | | Descriptor: | CADMIUM ION, Methyl Parathion Hydrolase, POTASSIUM ION, ... | | Authors: | Dong, Y, Sun, L, Bartlam, M, Rao, Z, Zhang, X. | | Deposit date: | 2003-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3
To be Published
|
|
1I85
 
 | | CRYSTAL STRUCTURE OF THE CTLA-4/B7-2 COMPLEX | | Descriptor: | CYTOTOXIC T-LYMPHOCYTE-ASSOCIATED PROTEIN 4, T LYMPHOCYTE ACTIVATION ANTIGEN CD86 | | Authors: | Schwartz, J.-C.D, Zhang, X, Fedorov, A.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2001-03-12 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for co-stimulation by the human CTLA-4/B7-2 complex.
Nature, 410, 2001
|
|
1ICC
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5 OUTER MITOCHONDRIAL MEMBRANE ISOFORM, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Terzyan, S, Zhang, X. | | Deposit date: | 2001-03-30 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the differences between rat liver outer mitochondrial membrane cytochrome b5 and microsomal cytochromes b5.
Biochemistry, 40, 2001
|
|
3KV5
 
 | | Structure of KIAA1718, human Jumonji demethylase, in complex with N-oxalylglycine | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV9
 
 | | Structure of KIAA1718 Jumonji domain | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, OXYGEN MOLECULE | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVA
 
 | | Structure of KIAA1718 Jumonji domain in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
