180L
 
 | |
172L
 
 | |
1R7R
 
 | | The crystal structure of murine p97/VCP at 3.6A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Huyton, T, Pye, V.E, Briggs, L.C, Flynn, T.C, Beuron, F, Kondo, H, Ma, J, Zhang, X, Freemont, P.S. | | Deposit date: | 2003-10-22 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of murine p97/VCP at 3.6A
J.Struct.Biol., 144, 2003
|
|
6CIJ
 
 | | Cryo-EM structure of mouse RAG1/2 HFC complex containing partial HMGB1 linker(3.9 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
4Q9V
 
 | | Crystal structure of TIPE3 | | Descriptor: | CHLORIDE ION, SULFATE ION, Tumor necrosis factor alpha-induced protein 8-like protein 3 | | Authors: | Wu, J, Zhang, X, Chen, Y.H, Shi, Y. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIPE3 Is the Transfer Protein of Lipid Second Messengers that Promote Cancer.
Cancer Cell, 26, 2014
|
|
4JPN
 
 | | Bacteriophage phiX174 H protein residues 143-221 | | Descriptor: | Minor spike protein H | | Authors: | Sun, L, Young, L.N, Boudko, S.B, Fokine, A, Zhang, X, Rossmann, M.G, Fane, B.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Icosahedral bacteriophage Phi X174 forms a tail for DNA transport during infection.
Nature, 505, 2014
|
|
4LDT
 
 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBCH5B~Ub | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ubiquitin, ... | | Authors: | Wiener, R, DiBello, A.T, Lombardi, P.M, Guzzo, C.M, Zhang, X, Matunis, M.J, Wolberger, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1FLC
 
 | | X-RAY STRUCTURE OF THE HAEMAGGLUTININ-ESTERASE-FUSION GLYCOPROTEIN OF INFLUENZA C VIRUS | | Descriptor: | HAEMAGGLUTININ-ESTERASE-FUSION GLYCOPROTEIN, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rosenthal, P.B, Zhang, X, Formanowski, F, Fitz, W, Wong, C.H, Meier-Ewert, H, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1999-02-22 | | Release date: | 2000-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus.
Nature, 396, 1998
|
|
6CG0
 
 | | Cryo-EM structure of mouse RAG1/2 HFC complex (3.17 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
3KVB
 
 | | Structure of KIAA1718 Jumonji domain in complex with N-oxalylglycine | | Descriptor: | JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6A96
 
 | | Cryo-EM structure of the human alpha5beta3 GABAA receptor in complex with GABA and Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-5,Gamma-aminobutyric acid receptor subunit alpha-5, ... | | Authors: | Liu, S, Xu, L, Guan, F, Liu, Y.T, Cui, Y, Zhang, Q, Bi, G.Q, Zhou, Z.H, Zhang, X, Ye, S. | | Deposit date: | 2018-07-11 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM structure of the human alpha 5 beta 3 GABAAreceptor.
Cell Res., 28, 2018
|
|
3O05
 
 | | Crystal Structure of Yeast Pyridoxal 5-Phosphate Synthase Snz1 Complxed with Substrate PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | Authors: | Teng, Y.B, Zhang, X, He, Y.X, Hu, H.X, Zhou, C.Z. | | Deposit date: | 2010-07-19 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
3O06
 
 | |
1TU4
 
 | | Crystal Structure of Rab5-GDP Complex | | Descriptor: | COBALT (II) ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-5A, ... | | Authors: | Zhu, G, Zhai, P, Liu, J, Terzyan, S, Li, G, Zhang, X.C. | | Deposit date: | 2004-06-24 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of Rab5-Rabaptin5 interaction in endocytosis
Nat.Struct.Mol.Biol., 11, 2004
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
4M8M
 
 | |
3O07
 
 | | Crystal structure of yeast pyridoxal 5-phosphate synthase Snz1 complexed with substrate G3P | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | Authors: | Teng, Y.B, Zhang, X, Hu, H.X, Zhou, C.Z. | | Deposit date: | 2010-07-19 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
1KHC
 
 | | Crystal Structure of the PWWP Domain of Mammalian DNA Methyltransferase Dnmt3b | | Descriptor: | DNA cytosine-5 methyltransferase 3B2, UNKNOWN ATOM OR ION | | Authors: | Qiu, C, Sawada, K, Zhang, X, Cheng, X. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds.
Nat.Struct.Biol., 9, 2002
|
|
178L
 
 | | Protein flexibility and adaptability seen in 25 crystal forms of T4 LYSOZYME | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Matsumura, M, Weaver, L, Zhang, X.-J, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
3WY7
 
 | | Crystal structure of Mycobacterium smegmatis 7-Keto-8-aminopelargonic acid (KAPA) synthase BioF | | Descriptor: | 8-amino-7-oxononanoate synthase | | Authors: | Fan, S.H, Li, D.F, Wang, D.C, Chen, G.J, Zhang, X.E, Bi, L.J. | | Deposit date: | 2014-08-20 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of Mycobacterium smegmatis 7-keto-8-aminopelargonic acid (KAPA) synthase
Int.J.Biochem.Cell Biol., 58C, 2014
|
|
5CP7
 
 | | Crystal Structure of an Antigen-Binding Fragment of Monoclonal Antibody against Sulfonamides | | Descriptor: | Heavy Chain of Antigen-Binding Fragment of Monoclonal Antibody of 4C7, Light Chain of Antigen-Binding Fragment of Monoclonal Antibody of 4C7 | | Authors: | Wang, Z, Shen, J, Li, C, Li, Y, Wen, K, Yu, X, Zhang, X. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Class-specific Monoclonal Antibodies and Dihydropteroate Synthase in Bioassays used for the Detection of Sulfonamides: Structural Insights into Recognition Diversity.
Anal. Chem., 91, 2019
|
|
3GT8
 
 | | Crystal structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jura, N, Endres, N.F, Engel, K, Deindl, S, Das, R, Lamers, M.H, Wemmer, D.E, Zhang, X, Kuriyan, J. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment.
Cell(Cambridge,Mass.), 137, 2009
|
|
5CP3
 
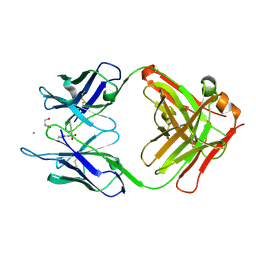 | | Crystal Structure of an Antigen-Binding Fragment of Monoclonal Antibody against Sulfonamides in Complex with Sulfathiazole | | Descriptor: | 4-amino-N-(1,3-thiazol-2-yl)benzenesulfonamide, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Z, Shen, J, Li, C, Li, Y, Wen, K, Yu, X, Zhang, X. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Class-specific Monoclonal Antibodies and Dihydropteroate Synthase in Bioassays used for the Detection of Sulfonamides: Structural Insights into Recognition Diversity.
Anal. Chem., 91, 2019
|
|
5Y9W
 
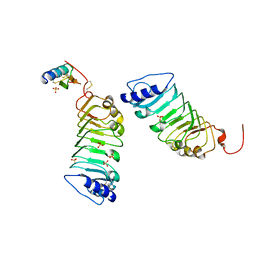 | | Crystal 1 for AtLURE1.2-AtPRK6LRR | | Descriptor: | Pollen receptor-like kinase 6, Protein LURE 1.2, SULFATE ION | | Authors: | Chai, J, Zhang, X. | | Deposit date: | 2017-08-28 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structural basis for receptor recognition of pollen tube attraction peptides.
Nat Commun, 8, 2017
|
|
