5ZGC
 
 | |
5BQX
 
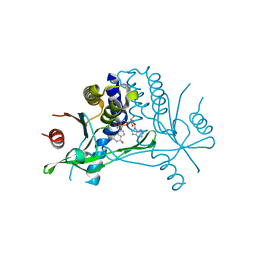 | | Crystal structure of human STING in complex with 3'2'-cGAMP | | Descriptor: | 3'2'-cGAMP, Stimulator of interferon genes protein | | Authors: | Wu, J, Zhang, X, Chen, Z.J, Chen, C. | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the specific recognition of the metazoan cyclic GMP-AMP by the innate immune adaptor protein STING.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5XNM
 
 | | Structure of unstacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
6IQL
 
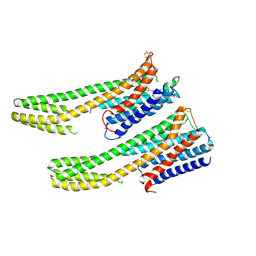 | | Crystal structure of dopamine receptor D4 bound to the subtype-selective ligand, L745870 | | Descriptor: | 3-{[4-(4-chlorophenyl)piperazin-1-yl]methyl}-1H-pyrrolo[2,3-b]pyridine, D(4) dopamine receptor,Soluble cytochrome b562,D(4) dopamine receptor | | Authors: | Zhou, Y, Cao, C, Zhang, X.C. | | Deposit date: | 2018-11-08 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of dopamine receptor D4 bound to the subtype selective ligand, L745870.
Elife, 8, 2019
|
|
6IZJ
 
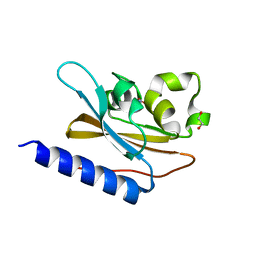 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, NreA | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2018-12-19 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
6K2H
 
 | | structural characterization of mutated NreA protein in nitrate binding site from staphylococcus aureus. | | Descriptor: | 1,2-ETHANEDIOL, NreA | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
7XKG
 
 | | Crystal structure of an intramolecular mesacyl-CoA transferase from the 3-hydroxypropionic acid cycle of Roseiflexus castenholzii | | Descriptor: | Acyl-CoA transferase/carnitine dehydratase-like protein | | Authors: | Min, Z.Z, Fan, C.P, Wu, W.P, Xin, Y.Y, Liu, M.H, Zhang, X, Wang, Z.G, Xu, X.L. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Intramolecular Mesaconyl-Coenzyme A Transferase From the 3-Hydroxypropionic Acid Cycle of Roseiflexus castenholzii .
Front Microbiol, 13, 2022
|
|
7DYB
 
 | | Thermotoga maritima ferritin mutant-FLAL-L | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Zhao, G, Zhang, X. | | Deposit date: | 2021-01-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials.
Nat Commun, 12, 2021
|
|
7D3U
 
 | | Structure of Mrp complex from Dietzia sp. DQ12-45-1b | | Descriptor: | Cation antiporter, DODECYL-BETA-D-MALTOSIDE, Monovalent Na+/H+ antiporter subunit A, ... | | Authors: | Li, B, Zhang, K.D, Wu, X.L, Zhang, X.C. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Dietzia Mrp complex reveals molecular mechanism of this giant bacterial sodium proton pump.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KHJ
 
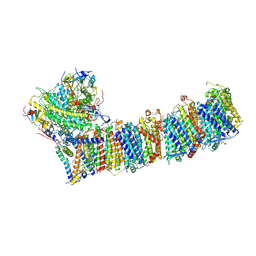 | | Supercomplex for electron transfer | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
5ZKQ
 
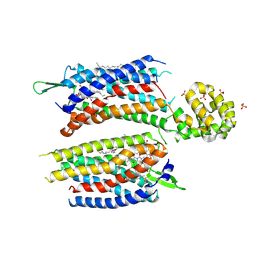 | | Crystal structure of the human platelet-activating factor receptor in complex with ABT-491 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-ethynyl-3-{3-fluoro-4-[(2-methyl-1H-imidazo[4,5-c]pyridin-1-yl)methyl]benzene-1-carbonyl}-N,N-dimethyl-1H-indole-1-carboxamide, Platelet-activating factor receptor,Endolysin,Endolysin,Platelet-activating factor receptor, ... | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8I4F
 
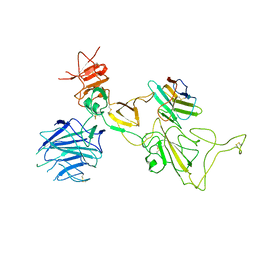 | | Omicron spike variant XBB with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4G
 
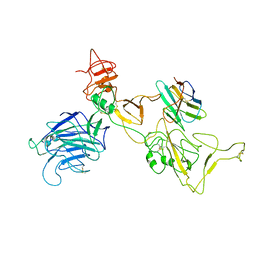 | | Omicron spike variant BQ.1.1 with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4E
 
 | | Omicron spike variant XBB with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4H
 
 | | Omicron spike variant BA.1 with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
5ZJI
 
 | | Structure of photosystem I supercomplex with light-harvesting complexes I and II | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Ma, J, Su, X.D, Cao, P, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-20 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II.
Science, 360, 2018
|
|
5ZKP
 
 | | Crystal structure of the human platelet-activating factor receptor in complex with SR 27417 | | Descriptor: | FLAVIN MONONUCLEOTIDE, N1,N1-dimethyl-N2-[(pyridin-3-yl)methyl]-N2-{4-[2,4,6-tri(propan-2-yl)phenyl]-1,3-thiazol-2-yl}ethane-1,2-diamine, Platelet-activating factor receptor,Flavodoxin,Platelet-activating factor receptor | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8WQL
 
 | | In situ PBS-PSII supercomplex from cyanobacterial Spirulina platensis | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | You, X, Zhang, X, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2023-10-11 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of in situ PBS-PSII supercomplex at 3.5 Angstroms resolution.
To Be Published
|
|
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
6AH0
 
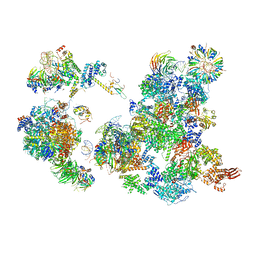 | | The Cryo-EM Structure of the Precusor of Human Pre-catalytic Spliceosome (pre-B complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
5XEX
 
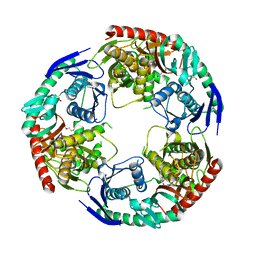 | | Crystal structure of S.aureus PNPase catalytic domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PYROPHOSPHATE, ... | | Authors: | Wang, X, Zhang, X, Zang, J. | | Deposit date: | 2017-04-06 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enolase binds to RnpA in competition with PNPase in Staphylococcus aureus
FEBS Lett., 591, 2017
|
|
6AHD
 
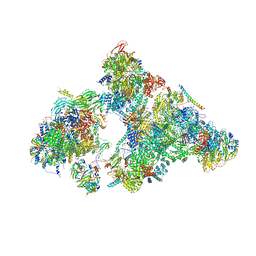 | | The Cryo-EM Structure of Human Pre-catalytic Spliceosome (B complex) at 3.8 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Brr2, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-14 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
6IJO
 
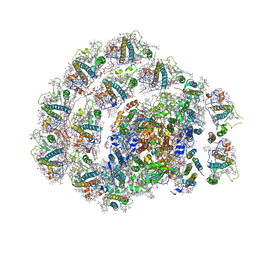 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
7CFP
 
 | |
7CFQ
 
 | | Crystal structure of WDR5 in complex with H3K4me3Q5ser peptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, H3K4me3Q5ser peptide, ... | | Authors: | Zhao, J, Zhang, X, Zang, J. | | Deposit date: | 2020-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the recognition of histone H3Q5 serotonylation by WDR5.
Sci Adv, 7, 2021
|
|
