6R9G
 
 | |
3UT8
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | 分子名称: | IODIDE ION, Putative uncharacterized protein | | 著者 | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | 登録日 | 2011-11-25 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.168 Å) | | 主引用文献 | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3UT4
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | 分子名称: | Putative uncharacterized protein | | 著者 | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | 登録日 | 2011-11-25 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
2L6K
 
 | | Solution Structure of a Nonphosphorylated Peptide Recognizing Domain | | 分子名称: | Tensin-like C1 domain-containing phosphatase | | 著者 | Dai, K, Liao, S, Zhang, J, Zhang, X, Tu, X. | | 登録日 | 2010-11-22 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Tensin2 SH2 domain and its phosphotyrosine-independent interaction with DLC-1
Plos One, 6, 2011
|
|
4FNC
 
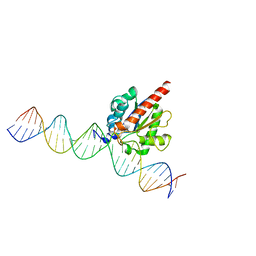 | | Human TDG in a post-reactive complex with 5-hydroxymethyluracil (5hmU) | | 分子名称: | 5-HYDROXYMETHYL URACIL, DNA (28-MER), DNA (29-MER), ... | | 著者 | Hashimoto, H, Hong, S, Bhagwat, A.S, Zhang, X, Cheng, X. | | 登録日 | 2012-06-19 | | 公開日 | 2012-09-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.493 Å) | | 主引用文献 | Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
3CI9
 
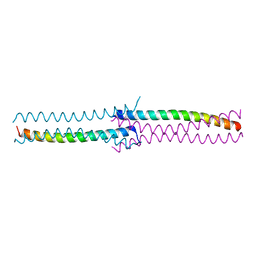 | | Crystal Structure of the human HSBP1 | | 分子名称: | Heat shock factor-binding protein 1 | | 著者 | Liu, X, Xu, L, Liu, Y, Zhu, G, Zhang, X.C, Li, X, Rao, Z. | | 登録日 | 2008-03-11 | | 公開日 | 2009-01-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the hexamer of human heat shock factor binding protein 1
Proteins, 75, 2009
|
|
3BU7
 
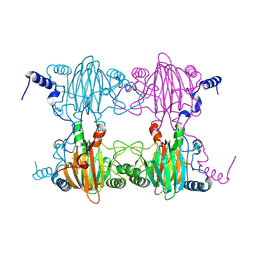 | | Crystal Structure and Biochemical Characterization of GDOsp, a Gentisate 1,2-Dioxygenase from Silicibacter Pomeroyi | | 分子名称: | FE (II) ION, Gentisate 1,2-dioxygenase | | 著者 | Chen, J, Wang, M.Z, Zhu, G.Y, Zhang, X.C, Rao, Z.H. | | 登録日 | 2008-01-02 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure and mutagenic analysis of GDOsp, a gentisate 1,2-dioxygenase from Silicibacter pomeroyi.
Protein Sci., 17, 2008
|
|
6LJS
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-[(2-phenylphenyl)amino]benzoic acid, Fatty acid-binding protein, ... | | 著者 | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | 登録日 | 2019-12-17 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6K5H
 
 | |
4ZDS
 
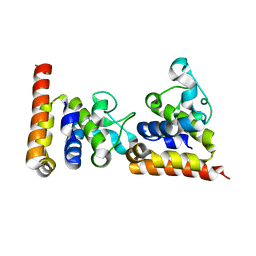 | | Crystal Structure of core DNA binding domain of Arabidopsis Thaliana Transcription Factor Ethylene-Insensitive 3 | | 分子名称: | Protein ETHYLENE INSENSITIVE 3 | | 著者 | Song, J, Zhu, C, Zhang, X, Wen, X, Liu, L, Peng, J, Guo, H, Yi, C. | | 登録日 | 2015-04-18 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Biochemical and Structural Insights into the Mechanism of DNA Recognition by Arabidopsis ETHYLENE INSENSITIVE3.
Plos One, 10, 2015
|
|
7D3U
 
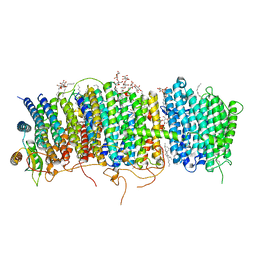 | | Structure of Mrp complex from Dietzia sp. DQ12-45-1b | | 分子名称: | Cation antiporter, DODECYL-BETA-D-MALTOSIDE, Monovalent Na+/H+ antiporter subunit A, ... | | 著者 | Li, B, Zhang, K.D, Wu, X.L, Zhang, X.C. | | 登録日 | 2020-09-21 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the Dietzia Mrp complex reveals molecular mechanism of this giant bacterial sodium proton pump.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | 分子名称: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | 著者 | Bhabha, G, Zhang, X, Ekiert, D.C. | | 登録日 | 2014-02-14 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6D2K
 
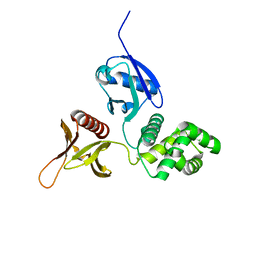 | |
6X19
 
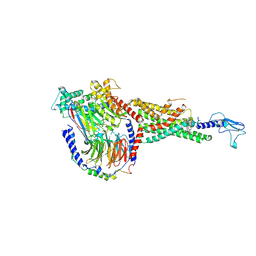 | | Non peptide agonist CHU-128, bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | 分子名称: | 3-{(1S)-1-[5-(2,2-dimethylmorpholin-4-yl)-2-{(4S)-2-(4-fluoro-3,5-dimethylphenyl)-3-[3-(4-fluoro-1-methyl-1H-indazol-5-yl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl]-4-methyl-2,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carbonyl}-1H-indol-1-yl]butyl}-1,2,4-oxadiazol-5(4H)-one, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Belousoff, M.J, Zhang, X, Danev, R. | | 登録日 | 2020-05-18 | | 公開日 | 2020-09-09 | | 最終更新日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
6X18
 
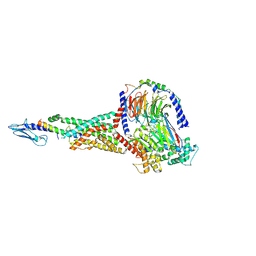 | | GLP-1 peptide hormone bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | 分子名称: | Glucagon, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Belousoff, M.J, Zhang, X, Danev, R. | | 登録日 | 2020-05-18 | | 公開日 | 2020-09-09 | | 最終更新日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
6KHI
 
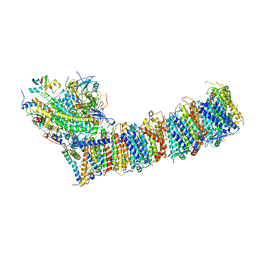 | | Supercomplex for cylic electron transport in cyanobacteria | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | 登録日 | 2019-07-15 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
3L64
 
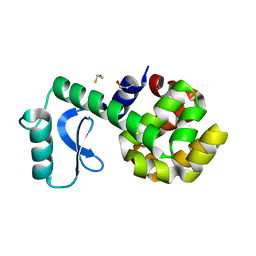 | | T4 Lysozyme S44E/WT* | | 分子名称: | BETA-MERCAPTOETHANOL, Lysozyme | | 著者 | Blaber, M, Zhang, X.-J, Lindstrom, J.D, Pepiot, S.D, Baase, W.A, Matthews, B.W. | | 登録日 | 2009-12-23 | | 公開日 | 2010-01-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Determination of alpha-helix propensity within the context of a folded protein. Sites 44 and 131 in bacteriophage T4 lysozyme.
J.Mol.Biol., 235, 1994
|
|
6KIF
 
 | | Structure of cyanobacterial photosystem I-IsiA-flavodoxin supercomplex | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | 登録日 | 2019-07-18 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-03-04 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
3KVB
 
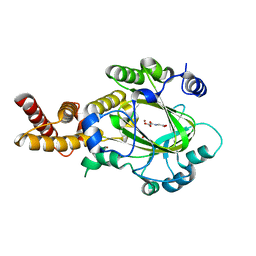 | | Structure of KIAA1718 Jumonji domain in complex with N-oxalylglycine | | 分子名称: | JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVA
 
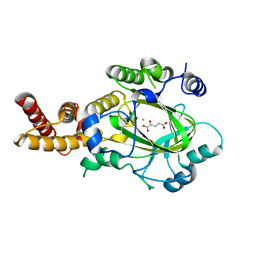 | | Structure of KIAA1718 Jumonji domain in complex with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV6
 
 | | Structure of KIAA1718, human Jumonji demethylase, in complex with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5NSS
 
 | | Cryo-EM structure of RNA polymerase-sigma54 holoenzyme with promoter DNA and transcription activator PspF intermedate complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta',DNA-directed RNA polymerase subunit beta',DNA-directed RNA polymerase subunit beta', ... | | 著者 | Glyde, R, Ye, F.Z, Darbari, V.C, Zhang, N, Buck, M, Zhang, X.D. | | 登録日 | 2017-04-26 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Structures of RNA Polymerase Closed and Intermediate Complexes Reveal Mechanisms of DNA Opening and Transcription Initiation.
Mol. Cell, 67, 2017
|
|
5NSR
 
 | | Cryo-EM structure of RNA polymerase-sigma54 holo enzyme with promoter DNA closed complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Glyde, R, Ye, F.Z, Darbari, V.C, Zhang, N, Buck, M, Zhang, X.D. | | 登録日 | 2017-04-26 | | 公開日 | 2017-06-28 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structures of RNA Polymerase Closed and Intermediate Complexes Reveal Mechanisms of DNA Opening and Transcription Initiation.
Mol. Cell, 67, 2017
|
|
3KXI
 
 | | crystal structure of SsGBP and GDP complex | | 分子名称: | GTP-binding protein (HflX), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Huang, B, Li, X, Zhang, X.C, Rao, Z. | | 登録日 | 2009-12-03 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Functional study on GTP hydrolysis by the GTP binding protein from Sulfolobus solfataricus, a member of the HflX family.
J.Biochem., 2010
|
|
3KXK
 
 | |
