3K18
 
 | | Tellurium modified DNA-8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(TTI)P*AP*CP*AP*C)-3' | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Huang, Z. | | Deposit date: | 2009-09-25 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and Structure Study of Oligonucleotides Derivatized with Tellurium Functionality
To be Published
|
|
8IQ6
 
 | | Cryo-EM structure of Latanoprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
8IQ4
 
 | | Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
6CV0
 
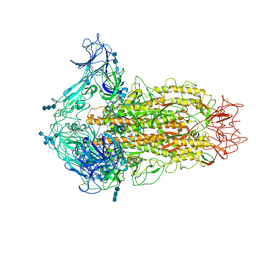 | | Cryo-electron microscopy structure of infectious bronchitis coronavirus spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Zheng, Y, Yang, Y, Liu, C, Geng, Q, Luo, C, Zhang, W, Li, F. | | Deposit date: | 2018-03-27 | | Release date: | 2018-04-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins.
PLoS Pathog., 14, 2018
|
|
3NSS
 
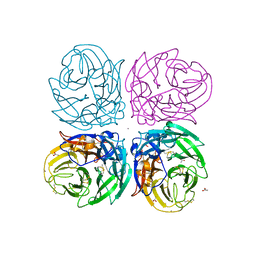 | | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active sites | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J.X, Zhang, W, Vavricka, C.J, Shi, Y, Gao, G.F. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site
Nat.Struct.Mol.Biol., 17, 2010
|
|
3ORR
 
 | | Crystal Structure of N5-Carboxyaminoimidazole synthetase from Staphylococcus aureus | | Descriptor: | N5-carboxyaminoimidazole ribonucleotide synthetase | | Authors: | Brugarolas, P, Duguid, E.M, Zhang, W, Poor, C.B, He, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3ORQ
 
 | | Crystal Structure of N5-Carboxyaminoimidazole synthetase from Staphylococcus aureus complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brugarolas, P, Duguid, E.M, Zhang, W, Poor, C.B, He, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-20 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4JFG
 
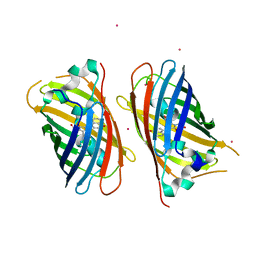 | | Crystal structure of sfGFP-66-HqAla | | Descriptor: | CESIUM ION, Green fluorescent protein, quinolin-8-ol | | Authors: | Wang, J, Liu, X, Li, J, Zhang, W, Hu, M, Zhou, J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Significant expansion of the fluorescent protein chromophore through the genetic incorporation of a metal-chelating unnatural amino acid.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3ORS
 
 | | Crystal Structure of N5-Carboxyaminoimidazole Ribonucleotide Mutase from Staphylococcus aureus | | Descriptor: | N5-Carboxyaminoimidazole Ribonucleotide Mutase, SULFATE ION | | Authors: | Brugarolas, P, Duguid, E.M, Zhang, W, Poor, C.B, He, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4LUQ
 
 | |
7L97
 
 | | Crystal structure of STAMBPL1 in complex with an engineered binder | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, SULFATE ION, ... | | Authors: | Guo, Y, Dong, A, Hou, F, Li, Y, Zhang, W, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of ubiquitin variant inhibitors for the JAMM-family deubiquitinases STAMBP and STAMBPL1.
J.Biol.Chem., 297, 2021
|
|
4QY0
 
 | | Structure of H10 from human-infecting H10N8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4QY2
 
 | | Structure of H10 from human-infecting H10N8 virus in complex with human receptor analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4FQB
 
 | |
5MDN
 
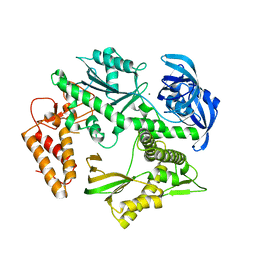 | | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis | | Descriptor: | DNA polymerase, MAGNESIUM ION | | Authors: | Guo, J, Zhang, W, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Akhtar, M. | | Deposit date: | 2016-11-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4FQA
 
 | |
2L89
 
 | |
4HTL
 
 | | Lmo2764 protein, a putative N-acetylmannosamine kinase, from Listeria monocytogenes | | Descriptor: | 1,2-ETHANEDIOL, Beta-glucoside kinase | | Authors: | Osipiuk, J, Mack, J, Endres, M, Salazar, J, Zhang, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-01 | | Release date: | 2012-11-14 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Lmo2764 protein, a putative N-acetylmannosamine kinase, from Listeria monocytogenes.
To be Published
|
|
4HCK
 
 | | HUMAN HCK SH3 DOMAIN, NMR, 25 STRUCTURES | | Descriptor: | HEMATOPOIETIC CELL KINASE | | Authors: | Horita, D.A, Baldisseri, D.M, Zhang, W, Altieri, A.S, Smithgall, T.E, Gmeiner, W.H, Byrd, R.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human Hck SH3 domain and identification of its ligand binding site.
J.Mol.Biol., 278, 1998
|
|
7MOO
 
 | | L-type DNA containing 2'-fluoro-2'-deoxycytidine | | Descriptor: | DNA (5'-D(*(0DG)P*(OFC)P*(0DG)P*(0DT)P*(0DA)P*(0DC)P*(0DG)P*(0DC))-3') | | Authors: | Dantsu, Y, Zhang, Y, Zhang, W. | | Deposit date: | 2021-05-03 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Synthesis of 2'-Deoxy-2'-fluoro-L-cytidine and Fluorinated L-Nucleic Acids for Structural Studies
Chemistryselect, 6, 2021
|
|
3FHA
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, GLYCEROL, ... | | Authors: | Yin, J, Li, L, Shaw, N, Li, Y, Song, J.K, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.C, Wang, L.X, Wang, P, Liu, Z.J. | | Deposit date: | 2008-12-09 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A.
Plos One, 4, 2009
|
|
3FHQ
 
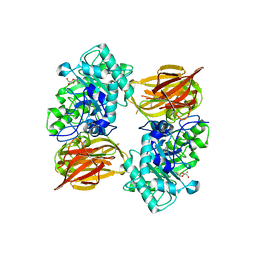 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Endo-beta-N-acetylglucosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | Authors: | Jie, Y, Li, L, Shaw, N, Li, Y, Song, J, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.-C, Wang, L.-X, Wang, P, Liu, Z.-J. | | Deposit date: | 2008-12-10 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A
Plos One, 4, 2009
|
|
7P6B
 
 | | Limbic-predominant neuronal inclusion body 4R tauopathy type 1b tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7Q4M
 
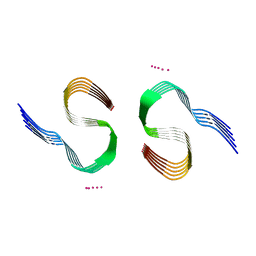 | | Type II beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-11-01 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
7Q4B
 
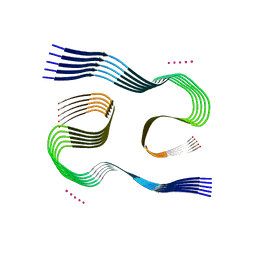 | | Type I beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-10-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
