4H5A
 
 | |
2L7E
 
 | |
4JUJ
 
 | | Crystal structure of 1918 pandemic influenza virus hemagglutinin mutant D225G complexed with human receptor analogue LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhang, W, Shi, Y, Qi, J, Gao, F, Li, Q, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution
J.Virol., 87, 2013
|
|
4I1G
 
 | |
4JUH
 
 | | Crystal structure of 1918 pandemic influenza virus hemagglutinin mutant D225G complexed with avian receptor analogue LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhang, W, Shi, Y, Qi, J, Gao, F, Li, Q, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-03-24 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution
J.Virol., 87, 2013
|
|
4K67
 
 | | Structure of an airborne transmissible avian influenza H5 hemagglutinin mutant from the influenza virus A/Indonesia/5/2005 complexed with human receptor analog LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose, ... | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
2L42
 
 | |
2L83
 
 | | A protein from Haloferax volcanii | | Descriptor: | Small archaeal modifier protein 1 | | Authors: | Zhang, W, Liao, S, Fan, K, Tu, X. | | Deposit date: | 2011-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
3E6U
 
 | | Crystal structure of Human LanCL1 | | Descriptor: | LanC-like protein 1, ZINC ION | | Authors: | Zhang, W, Zhu, G, Li, X, Rao, Z, Zhang, C. | | Deposit date: | 2008-08-16 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione
Genes Dev., 23, 2009
|
|
3E73
 
 | | Crystal Structure of Human LanCL1 complexed with GSH | | Descriptor: | GLUTATHIONE, LanC-like protein 1, ZINC ION | | Authors: | Zhang, W, Zhu, G, Li, X, Rao, Z, Zhang, C. | | Deposit date: | 2008-08-17 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione
Genes Dev., 23, 2009
|
|
5WCO
 
 | | Matrix Protein (M1) of Infectious Salmon Anaemia Virus | | Descriptor: | NS2 | | Authors: | Zhang, W, Zheng, W, Toh, Y, Betancourt, M.A, Tu, J, Fan, Y, Vakharia, V, Liu, J, McNew, J.A, Jin, M, Tao, Y.J. | | Deposit date: | 2017-07-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal structure of an orthomyxovirus matrix protein reveals mechanisms for self-polymerization and membrane association.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ZHY
 
 | | Structural characterization of the HCoV-229E fusion core | | Descriptor: | Spike glycoprotein | | Authors: | Zhang, W, Zheng, Q, Yan, M, Chen, X, Yang, H, Zhou, W, Rao, Z. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.441 Å) | | Cite: | Structural characterization of the HCoV-229E fusion core.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
5ZHZ
 
 | | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis | | Descriptor: | Probable endonuclease 4, SULFATE ION, ZINC ION | | Authors: | Zhang, W, Xu, Y, Yan, M, Li, S, Wang, H, Yang, H, Zhou, W, Rao, Z. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis.
Biochem. Biophys. Res. Commun., 498, 2018
|
|
6L15
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L12
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 4-[(2-chloranylphenoxazin-10-yl)methyl]cyclohexan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L16
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-[4-[2-(7-chloranylpyrido[3,4-b][1,4]benzoxazin-5-yl)ethyl]piperidin-1-yl]ethanamine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L17
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazin-8-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L11
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 5-(2-chloranylphenoxazin-10-yl)-~{N},~{N}-diethyl-pentan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L13
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-(2-piperidin-4-ylethyl)phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L14
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-[3-[(3~{S})-piperidin-3-yl]propyl]phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
3CMI
 
 | |
8SX6
 
 | | RNA duplex bound with GMP and AMP monomers | | Descriptor: | ADENOSINE MONOPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SWG
 
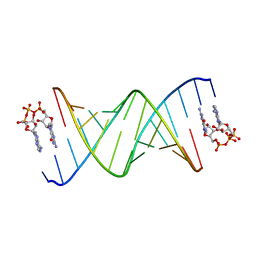 | | RNA duplex bound with GpppA dinucleotide ligand | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GA3))-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SX5
 
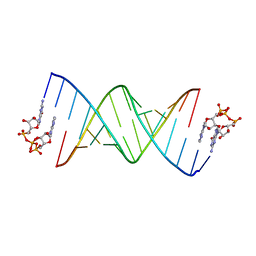 | | GpppA dinucleotide binding to RNA CU template | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA (5'-R(*(TLN)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GA3))-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SXL
 
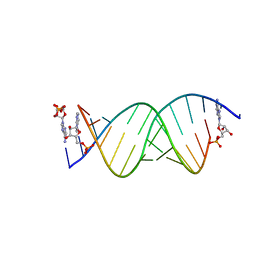 | | RNA UU template binding to AMP monomer | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA (5'-R(*(TLN)P*(TLN)P*(LCA)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-22 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
