3K32
 
 | |
7MR1
 
 | |
7MR0
 
 | |
7MR3
 
 | | Cryo-EM structure of RecBCD-DNA complex with docked RecBNuc and stabilized RecD | | Descriptor: | DNA (60-MER), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Hao, L, Zhang, R, Lohman, T.M. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Heterogeneity in E. coli RecBCD Helicase-DNA Binding and Base Pair Melting.
J.Mol.Biol., 433, 2021
|
|
7MR4
 
 | | Cryo-EM structure of RecBCD-DNA complex with undocked RecBNuc and flexible RecD | | Descriptor: | DNA (60-MER), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Hao, L, Zhang, R, Lohman, T.M. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Heterogeneity in E. coli RecBCD Helicase-DNA Binding and Base Pair Melting.
J.Mol.Biol., 433, 2021
|
|
7MR2
 
 | |
1XTN
 
 | | crystal structure of CISK-PX domain with sulfates | | Descriptor: | SULFATE ION, Serine/threonine-protein kinase Sgk3 | | Authors: | Xing, Y, Liu, D, Zhang, R, Joachimiak, A, Songyang, Z, Xu, W. | | Deposit date: | 2004-10-22 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX)
J.Biol.Chem., 279, 2004
|
|
1XTE
 
 | | crystal structure of CISK-PX domain | | Descriptor: | Serine/threonine-protein kinase Sgk3 | | Authors: | Xing, Y, Liu, D, Zhang, R, Joachimiak, A, Songyang, Z, Xu, W. | | Deposit date: | 2004-10-21 | | Release date: | 2004-11-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX)
J.Biol.Chem., 279, 2004
|
|
1XV2
 
 | | Crystal Structure of a Protein of Unknown Function Similar to Alpha-acetolactate Decarboxylase from Staphylococcus aureus | | Descriptor: | ZINC ION, hypothetical protein, similar to alpha-acetolactate decarboxylase | | Authors: | Duke, N.E.C, Zhang, R, Quartey, P, Collart, F, Holzle, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a hypothetical protein similar to alpha-acetolactate
To be Published
|
|
1Y7R
 
 | | 1.7 A Crystal Structure of Protein of Unknown Function SA2161 from Meticillin-Resistant Staphylococcus aureus, Probable Acetyltransferase | | Descriptor: | PHOSPHATE ION, hypothetical protein SA2161 | | Authors: | Qiu, Y, Zhang, R, Collart, F, Holzle, D, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-09 | | Release date: | 2005-01-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7A Crystal Structure of Hypothetical Protein SA2161 from Meticillin-Resistant Staphylococcus aureus
To be Published
|
|
1Y89
 
 | | Crystal Structure of devB protein | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DI(HYDROXYETHYL)ETHER, NONAETHYLENE GLYCOL, ... | | Authors: | Lazarski, K, Cymborowski, M, Chruszcz, M, Zheng, H, Zhang, R, Lezondra, L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of devB protein
To be Published
|
|
1YOZ
 
 | | Predicted coding region AF0941 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0941 | | Authors: | Lunin, V.V, Zhang, R, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-28 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of predicted coding region AF0941 from Archaeoglobus fulgidus
To be Published
|
|
1NRW
 
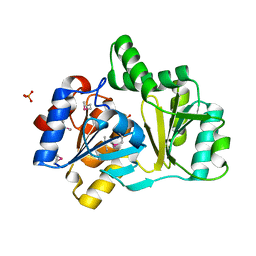 | | The structure of a HALOACID DEHALOGENASE-LIKE HYDROLASE FROM B. SUBTILIS | | Descriptor: | CALCIUM ION, PHOSPHATE ION, hypothetical protein, ... | | Authors: | Cuff, M.E, Kim, Y, Zhang, R, Joachimiak, A, Collart, F, Quartey, P, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-25 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a HALOACID DEHALOGENASE-LIKE HYDROLASE FROM B. SUBTILIS
To be Published
|
|
1P53
 
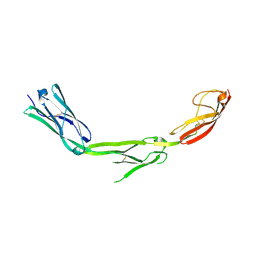 | | The Crystal Structure of ICAM-1 D3-D5 fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Intercellular adhesion molecule-1 | | Authors: | Yang, Y, Jun, C.D, Liu, J.H, Zhang, R, Jochimiak, A, Springer, T.A, Wang, J.H. | | Deposit date: | 2003-04-24 | | Release date: | 2004-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis for dimerization of ICAM-1 on the cell surface.
Mol.Cell, 14, 2004
|
|
3CM8
 
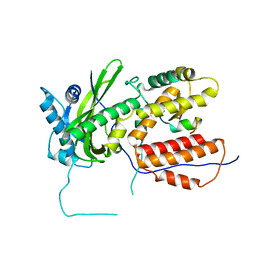 | | A RNA polymerase subunit structure from virus | | Descriptor: | Polymerase acidic protein, peptide from RNA-directed RNA polymerase catalytic subunit | | Authors: | He, X, Zhou, J, Zeng, Z, Ma, J, Zhang, R, Rao, Z, Liu, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2008-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Crystal structure of the polymerase PAC-PB1N complex from an avian influenza H5N1 virus
Nature, 454, 2008
|
|
1R4V
 
 | | 1.9A crystal structure of protein AQ328 from Aquifex aeolicus | | Descriptor: | CACODYLATE ION, Hypothetical protein AQ_328, ZINC ION | | Authors: | Qiu, Y, Tereshko, V, Kim, Y, Zhang, R, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-08 | | Release date: | 2004-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of Aq_328 from the hyperthermophilic bacteria Aquifex aeolicus shows an ancestral histone fold.
Proteins, 62, 2006
|
|
1ROC
 
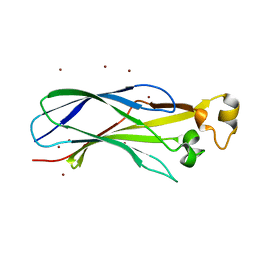 | | Crystal structure of the histone deposition protein Asf1 | | Descriptor: | Anti-silencing protein 1, BROMIDE ION | | Authors: | Daganzo, S.M, Erzberger, J.P, Lam, W.M, Skordalakes, E, Zhang, R, Franco, A.A, Brill, S.J, Adams, P.D, Berger, J.M, Kaufman, P.D. | | Deposit date: | 2003-12-02 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the conserved core of histone deposition protein Asf1.
Curr.Biol., 13, 2003
|
|
1SBX
 
 | | Crystal structure of the Dachshund-homology domain of human SKI | | Descriptor: | Ski oncogene | | Authors: | Wilson, J.J, Malakhova, M, Zhang, R, Joachimiak, A, Hegde, R.S. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Dachshund Homology Domain of human SKI
Structure, 12, 2004
|
|
1S9U
 
 | | Atomic structure of a putative anaerobic dehydrogenase component | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, putative component of anaerobic dehydrogenases | | Authors: | Qiu, Y, Zhang, R, Tereshko, V, Kim, Y, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-05 | | Release date: | 2004-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The 1.38 A crystal structure of DmsD protein from Salmonella typhimurium, a proofreading chaperone on the Tat pathway.
Proteins, 71, 2008
|
|
1T1J
 
 | | Crystal structure of genomics APC5043 | | Descriptor: | hypothetical protein | | Authors: | Dong, A, Xu, X, Liu, Y, Zhang, R, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein PA1492 from Pseudomonas aeruginosa
TO BE PUBLISHED
|
|
2QMW
 
 | | The crystal structure of the prephenate dehydratase (PDT) from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Zhang, R, Li, H, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-17 | | Release date: | 2007-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - implication of allosteric regulation by L-phenylalanine.
J.Struct.Biol., 162, 2008
|
|
4IR0
 
 | | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, FOSFOMYCIN, Metallothiol transferase FosB 2, ... | | Authors: | Maltseva, N, Kim, Y, Jedrzejczak, R, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames
To be Published
|
|
4DX9
 
 | | ICAP1 in complex with integrin beta 1 cytoplasmic tail | | Descriptor: | Integrin beta-1, Integrin beta-1-binding protein 1 | | Authors: | Liu, W, Draheim, K, Zhang, R, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-09 | | Last modified: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Mechanism for KRIT1 Release of ICAP1-Mediated Suppression of Integrin Activation.
Mol.Cell, 49, 2013
|
|
4DXA
 
 | | Co-crystal structure of Rap1 in complex with KRIT1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Krev interaction trapped protein 1, MAGNESIUM ION, ... | | Authors: | Li, X, Zhang, R, Boggon, T.J. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Small G Protein Effector Interaction of Ras-related Protein 1 (Rap1) and Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
J.Biol.Chem., 287, 2012
|
|
4DX8
 
 | | ICAP1 in complex with KRIT1 N-terminus | | Descriptor: | BROMIDE ION, Integrin beta-1-binding protein 1, Krev interaction trapped protein 1 | | Authors: | Liu, W, Draheim, K, Zhang, R, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Mechanism for KRIT1 Release of ICAP1-Mediated Suppression of Integrin Activation.
Mol.Cell, 49, 2013
|
|
