3SZ4
 
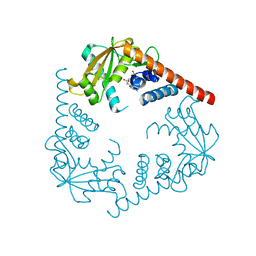 | | Crystal Structure of LHK-Exo in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
3SZ5
 
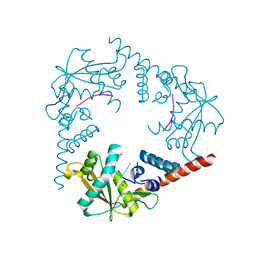 | | Crystal Structure of LHK-Exo in complex with 5-phosphorylated oligothymidine (dT)4 | | Descriptor: | 5'-D(P*TP*TP*TP*T)-3', Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
3QPS
 
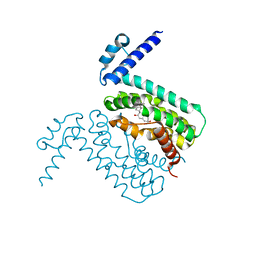 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
3QQA
 
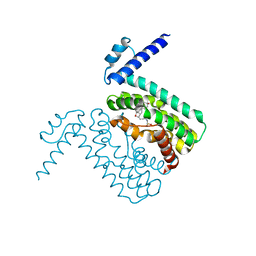 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.-C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
7V5H
 
 | |
7V9Z
 
 | |
7VA3
 
 | |
7VA6
 
 | |
7VA2
 
 | |
7X78
 
 | | L-fuculose 1-phosphate aldolase | | Descriptor: | L-fuculose phosphate aldolase, MAGNESIUM ION, SULFATE ION | | Authors: | Lou, X, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization of an L-fuculose-1-phosphate aldolase from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 607, 2022
|
|
8IJQ
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide | | Descriptor: | N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IJR
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with diacylglycerol/phosphoethanolamine | | Descriptor: | (2S)-1-(hexadecanoyloxy)-3-hydroxypropan-2-yl (11Z)-octadec-11-enoate, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7D54
 
 | | Crstal structure MsGATase with Gln | | Descriptor: | GLUTAMINE, Glutamine amidotransferase class-I | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D50
 
 | | SpuA mutant - H221N with glutamyl-thioester | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8GQV
 
 | | The Crystal Structures of a Swine SLA-2*HB01 Molecules Complexed with a CTL epitope from Asia1 serotype of Foot-and-mouth disease virus | | Descriptor: | As64, MHC class I antigen, beta 2 microglobulin | | Authors: | Feng, L, Gao, Y.Y, Sun, M.W, Li, Z.B, Zhang, Q, Yang, J, Qiao, C, Jin, H, Feng, H.S, Xian, Y.H, Qi, J.X, Gao, G.F, Liu, W.J, Gao, F.S. | | Deposit date: | 2022-08-31 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Parallel Presentation of Two Functional CTL Epitopes Derived from the O and Asia 1 Serotypes of Foot-and-Mouth Disease Virus and Swine SLA-2*HB01: Implications for Universal Vaccine Development.
Cells, 11, 2022
|
|
8GQW
 
 | | The Crystal Structures of a Swine SLA-2*HB01 Molecules Complexed with a CTL epitope from Asia1 serotype of Foot-and-mouth disease virus | | Descriptor: | Hu64, MHC class I antigen, beta 2 microglobulin | | Authors: | Feng, L, Gao, Y.Y, Sun, M.W, Li, Z.B, Zhang, Q, Yang, J, Qiao, C, Jin, H, Feng, H.S, Xian, Y.H, Qi, J.X, Gao, G.F, Liu, W.J, Gao, F.S. | | Deposit date: | 2022-08-31 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The Parallel Presentation of Two Functional CTL Epitopes Derived from the O and Asia 1 Serotypes of Foot-and-Mouth Disease Virus and Swine SLA-2*HB01: Implications for Universal Vaccine Development.
Cells, 11, 2022
|
|
8GPD
 
 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed penicillin V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxidanyl-2-oxidanylidene-1-(2-phenoxyethanoylamino)ethyl]-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, POTASSIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
8GPC
 
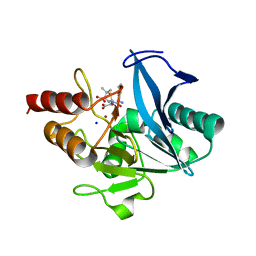 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, SODIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
8GPE
 
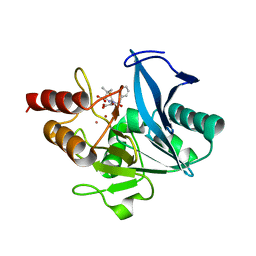 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed penicillin G | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, POTASSIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
7D4R
 
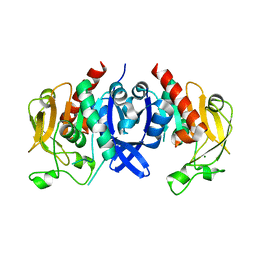 | | SpuA native structure | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D53
 
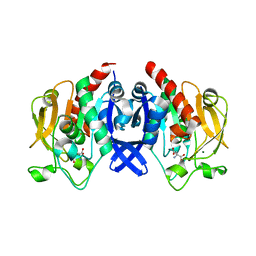 | | SpuA mutant - H221N with Glu | | Descriptor: | GLUTAMIC ACID, MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8Y85
 
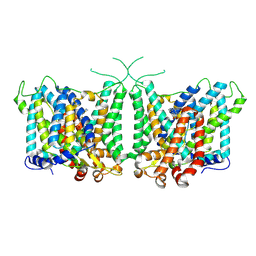 | | Human AE3 with NaHCO3- and DIDS | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Anion exchange protein 3, BICARBONATE ION | | Authors: | Jian, L, Zhang, Q, Yao, D, Wang, Q, Xia, Y, Qin, A, Cao, Y. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
8Y8K
 
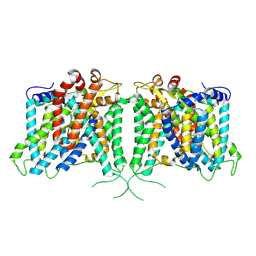 | | The structure of hAE3 | | Descriptor: | Anion exchange protein 3 | | Authors: | Jian, L, Zhang, Q, Yao, D, Wang, Q, Xia, Y, Qin, A, Cao, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
8ZLE
 
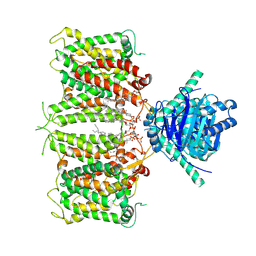 | | hAE3NTD2TMD with PT5,CLR, and Y01 | | Descriptor: | CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate, ... | | Authors: | Jian, L, Zhang, Q, Yao, D, Wang, Q, Xia, Y, Qin, A, Cao, Y. | | Deposit date: | 2024-05-19 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
7U4A
 
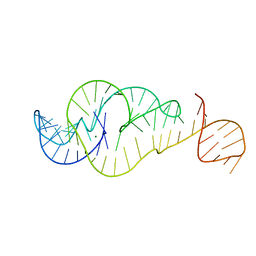 | | Crystal Structure of Zika virus xrRNA1 mutant | | Descriptor: | MAGNESIUM ION, RNA (70-MER) | | Authors: | Thompson, R.D, Carbaugh, D.L, Nielsen, J.R, Witt, C, Meganck, R.M, Rangadurai, A, Zhao, B, Bonin, J.P, Nathan, N.T, Marzluff, W.F, Frank, A.T, Lazear, H.M, Zhang, Q. | | Deposit date: | 2022-02-28 | | Release date: | 2023-09-06 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Lifetime of ground conformational state determines the activity of structured RNA.
Nat.Chem.Biol., 2025
|
|
