7N9R
 
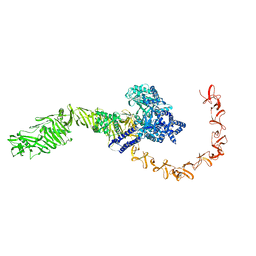 | | state 4 of TcdB and FZD2 at pH5 | | Descriptor: | Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N9S
 
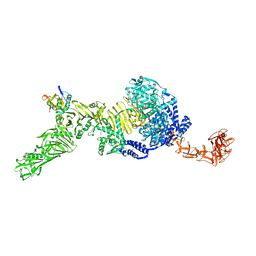 | | TcdB and frizzled-2 CRD complex | | Descriptor: | Frizzled-2, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N95
 
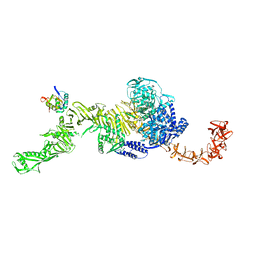 | | state 1 of TcdB and FZD2 at pH5 | | Descriptor: | Frizzled-2, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
3TKZ
 
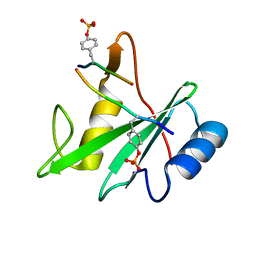 | | Structure of the SHP-2 N-SH2 domain in a 1:2 complex with RVIpYFVPLNR peptide | | Descriptor: | PROTEIN (RVIpYFVPLNR peptide), Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Y, Zhang, J, Yuan, C, Hard, R.L, Park, I.H, Li, C, Bell, C.E, Pei, D. | | Deposit date: | 2011-08-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain.
Biochemistry, 50, 2011
|
|
3TL0
 
 | | Structure of SHP2 N-SH2 domain in complex with RLNpYAQLWHR peptide | | Descriptor: | RLNpYAQLWHR peptide, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Y, Zhang, J, Yuan, C, Hard, R.L, Park, I.H, Li, C, Bell, C.E, Pei, D. | | Deposit date: | 2011-08-29 | | Release date: | 2011-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain.
Biochemistry, 50, 2011
|
|
8DSG
 
 | | P411-PFA carbene transferase | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P450-BM3 variant P411-PFA, ... | | Authors: | Maggiolo, A.O, Porter, N.J, Zhang, J, Arnold, F.H. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Chemodivergent C(sp 3 )-H and C(sp 2 )-H Cyanomethylation Using Engineered Carbene Transferases.
Nat Catal, 6, 2023
|
|
2I7K
 
 | | Solution Structure of the Bromodomain of Human BRD7 Protein | | Descriptor: | Bromodomain-containing protein 7 | | Authors: | Sun, H, Liu, J, Zhang, J, Huang, H, Wu, J, Shi, Y. | | Deposit date: | 2006-08-31 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4
Biochem.Biophys.Res.Commun., 358, 2007
|
|
7YRZ
 
 | | Crystal structure of HCoV 229E main protease in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Zhou, Y.R, Zeng, P, Zhou, X.L, Lin, C, Zhang, J, Yin, X.S, Li, J. | | Deposit date: | 2022-08-11 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis of main proteases of HCoV-229E bound to inhibitor PF-07304814 and PF-07321332.
Biochem.Biophys.Res.Commun., 657, 2023
|
|
2JXN
 
 | | Solution Structure of S. cerevisiae PDCD5-like Protein Ymr074cp | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, Uncharacterized protein YMR074C | | Authors: | Hong, J, Zhang, J, Liu, Z, Shi, Y, Wu, J. | | Deposit date: | 2007-11-23 | | Release date: | 2008-12-02 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of S. cerevisiae PDCD5-like Protein Ymr074cp Determined by Heteronuclear NMR Spectroscopy
To be Published
|
|
7XRY
 
 | | Crystal structure of MERS main protease in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2022-05-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7XRS
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Replicase polyprotein 1a | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-05-11 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7YGQ
 
 | | Crystal structure of SARS main protease in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-07-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7XB4
 
 | | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | Authors: | Hu, X.H, Li, J, Zhang, J. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for the Inhibition of SARS-CoV-2 M pro D48N Mutant by Shikonin and PF-07321332.
Viruses, 16, 2023
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
8H91
 
 | |
6VXN
 
 | | Cryo-EM structure of Arabidopsis thaliana MSL1 A320V | | Descriptor: | DODECANE, Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Deng, Z, Zhang, J, Yuan, P. | | Deposit date: | 2020-02-22 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural mechanism for gating of a eukaryotic mechanosensitive channel of small conductance.
Nat Commun, 11, 2020
|
|
6NM5
 
 | | F-pilus/MS2 Maturation protein complex | | Descriptor: | (2R)-2,3-dihydroxypropyl ethyl hydrogen (S)-phosphate, Maturation protein, Type IV conjugative transfer system pilin TraA | | Authors: | Meng, R, Chang, J, Zhang, J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural basis for the adsorption of a single-stranded RNA bacteriophage.
Nat Commun, 10, 2019
|
|
6VXM
 
 | | Cryo-EM structure of Arabidopsis thaliana MSL1 | | Descriptor: | EICOSANE, Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Deng, Z, Zhang, J, Yuan, P. | | Deposit date: | 2020-02-22 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural mechanism for gating of a eukaryotic mechanosensitive channel of small conductance.
Nat Commun, 11, 2020
|
|
6VXP
 
 | |
6ZRK
 
 | | Crystal structure of H8 haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Xiong, X, Walker, P, Zhang, J, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-07-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hemagglutinin Structure and Activities.
Cold Spring Harb Perspect Med, 11, 2021
|
|
6WDO
 
 | | Cryo-EM structure of mitochondrial calcium uniporter holocomplex in high Ca2+ | | Descriptor: | CALCIUM ION, Calcium uniporter protein, mitochondrial, ... | | Authors: | Feng, L, Zhang, J, Fan, M. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and mechanism of the mitochondrial Ca2+uniporter holocomplex.
Nature, 582, 2020
|
|
7A9D
 
 | | Crystal structure of H12 Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Xiong, X, Walker, P, Zhang, J, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hemagglutinin Structure and Activities.
Cold Spring Harb Perspect Med, 11, 2021
|
|
6WDN
 
 | | Cryo-EM structure of mitochondrial calcium uniporter holocomplex in low Ca2+ | | Descriptor: | Calcium uniporter protein, mitochondrial, Calcium uptake protein 1, ... | | Authors: | Feng, L, Zhang, J, Fan, M. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-27 | | Last modified: | 2020-06-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and mechanism of the mitochondrial Ca2+uniporter holocomplex.
Nature, 582, 2020
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
