7V4R
 
 | | The crystal structure of KFDV NS3H bound with Pi | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, Serine protease NS3 | | Authors: | Zhang, C.Y, Jin, T.C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kyasanur Forest disease virus NS3 helicase: Insights into structure, activity, and inhibitors.
Int.J.Biol.Macromol., 2023
|
|
7ESQ
 
 | |
7ESV
 
 | |
7ESP
 
 | |
7ESO
 
 | |
6L7O
 
 | | cryo-EM structure of cyanobacteria Fd-NDH-1L complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Zhang, C, Shuai, J, Wu, J, Lei, M. | | Deposit date: | 2019-11-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into NDH-1 mediated cyclic electron transfer.
Nat Commun, 11, 2020
|
|
8GH5
 
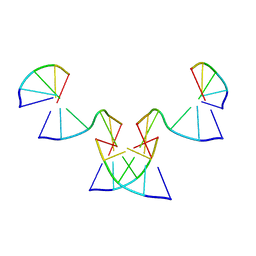 | | Implementing Logic Gates in DNA Crystal Engineering | | Descriptor: | DNA (5'-D(*AP*GP*AP*CP*G)-3'), DNA (5'-D(*CP*TP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*A)-3'), ... | | Authors: | Zhang, C, Paluzzi, V.E, Sha, R, Jonoska, N, Mao, C. | | Deposit date: | 2023-03-09 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Implementing Logic Gates by DNA Crystal Engineering.
Adv Mater, 35, 2023
|
|
7DCD
 
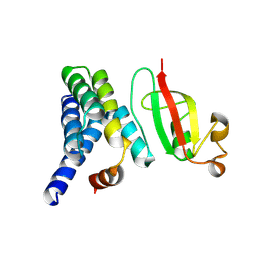 | |
9J6O
 
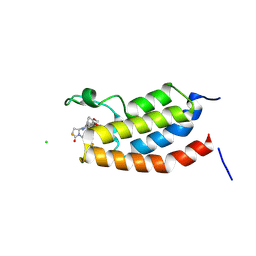 | | Crystal Structure of bromodomain of human CBP in complex with the inhibitor CZL-046 | | Descriptor: | (3~{S},5~{S})-1-[3,4-bis(fluoranyl)phenyl]-5-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]-3-fluoranyl-pyrrolidin-2-one, CHLORIDE ION, CREB-binding protein | | Authors: | Zhang, Y, Zhang, C, Chen, Z, Yang, H, Huang, X, Li, Y, Xu, Y. | | Deposit date: | 2024-08-16 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of CZL-046 with an ( S )-3-Fluoropyrrolidin-2-one Scaffold as a p300 Bromodomain Inhibitor for the Treatment of Multiple Myeloma.
J.Med.Chem., 2024
|
|
9JE1
 
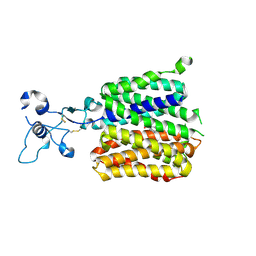 | | Human URAT1 bound to dotinurad | | Descriptor: | Solute carrier family 22 member 12, dotinurad | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
9JDY
 
 | | Human URAT1 bound with verinurad | | Descriptor: | Solute carrier family 22 member 12, verinurad | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
4XV2
 
 | |
4XV3
 
 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX7922 | | Descriptor: | N'-{3-[5-(2-aminopyrimidin-4-yl)-2-tert-butyl-1,3-thiazol-4-yl]-2-fluorophenyl}-N-ethyl-N-methylsulfuric diamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
4XV9
 
 | | B-Raf Kinase domain in complex with PLX5568 | | Descriptor: | N-{3-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]-2,4-difluorophenyl}-4-(trifluoromethyl)benzenesulfonamide, SULFATE ION, Serine/threonine-protein kinase B-raf | | Authors: | zhang, Y, zhang, c, wang, w. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
9JE0
 
 | | Human URAT1 bound to benzbromarone | | Descriptor: | Solute carrier family 22 member 12, [3,5-bis(bromanyl)-4-oxidanyl-phenyl]-(2-ethyl-1-benzofuran-3-yl)methanone | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
9JDV
 
 | | Human URAT1 bound with Uric acid | | Descriptor: | Solute carrier family 22 member 12, URIC ACID | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
7M8M
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 11 | | Descriptor: | 3C-like proteinase, 5-[3-(3-chloro-5-propoxyphenyl)-2-oxo-2H-[1,3'-bipyridin]-5-yl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8O
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 19 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(3-fluorophenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8P
 
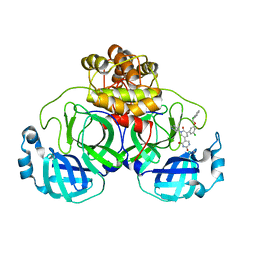 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 23 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8N
 
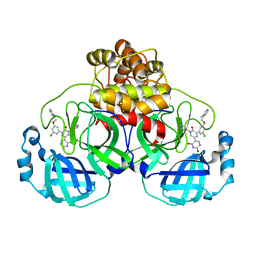 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 16 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(2-methylphenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8Y
 
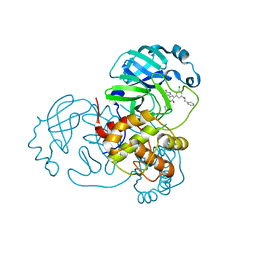 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 15 | | Descriptor: | 3C-like proteinase, 5-{3-[3-chloro-5-(2-phenylethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M3E
 
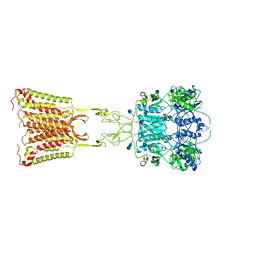 | | Asymmetric Activation of the Calcium Sensing Receptor Homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, ... | | Authors: | Gao, Y, Robertson, M.J, Zhang, C, Meyerowitz, J.G, Panova, O, Skiniotis, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Asymmetric activation of the calcium-sensing receptor homodimer.
Nature, 595, 2021
|
|
7M90
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 50 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[2-(3-oxopiperazin-1-yl)ethoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M3J
 
 | | Asymmetric Activation of the Calcium Sensing Receptor Homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, ... | | Authors: | Gao, Y, Robertson, M.J, Zhang, C, Meyerowitz, J.G, Panova, O, Skiniotis, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric activation of the calcium-sensing receptor homodimer.
Nature, 595, 2021
|
|
7M91
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 25 | | Descriptor: | 3C-like proteinase, 5-{3-[3-chloro-5-(3,3,3-trifluoropropoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
