8W6N
 
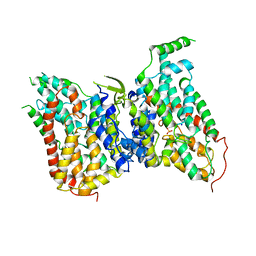 | | NaS1 with sulfate in IN/OUT state | | Descriptor: | SODIUM ION, SULFATE ION, Solute carrier family 13 member 1 | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-29 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6D
 
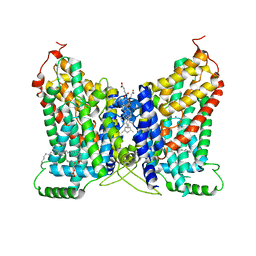 | | CryoEM structure of NaDC1 in apo state | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6O
 
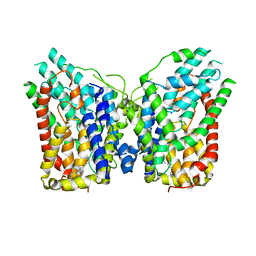 | | NaS1 in IN/IN state | | Descriptor: | CHOLESTEROL, SODIUM ION, Solute carrier family 13 member 1 | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-29 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6G
 
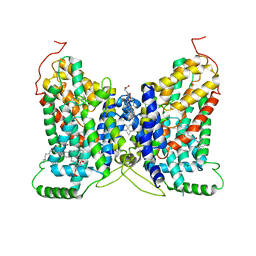 | | NaDC1 with inhibitor ACA | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-[[(~{E})-3-(4-pentylphenyl)prop-2-enoyl]amino]benzoic acid, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6T
 
 | | NaS1 in IN/OUT state | | Descriptor: | Solute carrier family 13 member 1 | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-29 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
6IVQ
 
 | | Crystal structure of a membrane protein S19A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVK
 
 | | Crystal structure of a membrane protein G175A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
8TWP
 
 | | Influenza A virus (A/Aichi/2/1968(H3N2) nucleoprotein mutant - 2-7 deleted, R416A | | Descriptor: | Nucleoprotein | | Authors: | Yoon, J, Zhang, Y.M, Grant, R.A, Shoulders, M.D. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The immune-evasive proline-283 substitution in influenza nucleoprotein increases aggregation propensity without altering the native structure.
Sci Adv, 10, 2024
|
|
6IVR
 
 | | Crystal structure of a membrane protein W16A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVP
 
 | | Crystal structure of a membrane protein P262A | | Descriptor: | CHLORIDE ION, ZINC ION, bestrophin | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVN
 
 | | Crystal structure of a membrane protein G264A | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVL
 
 | | Crystal structure of a membrane protein L259A | | Descriptor: | ACETIC ACID, CHLORIDE ION, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVJ
 
 | | Crystal structure of a membrane protein G18A | | Descriptor: | ACETIC ACID, CHLORIDE ION, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
8HYJ
 
 | |
8WA1
 
 | |
8WA0
 
 | |
8VQ4
 
 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-125A. | | Descriptor: | (8R)-6-(1-benzyl-1H-pyrazole-4-carbonyl)-N-[(2S,3R)-3-(2-cyclohexylethoxy)-1-(methylamino)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expanding the ligandable proteome by paralog hopping with covalent probes.
Biorxiv, 2024
|
|
8WTZ
 
 | | potassium outward rectifier channel SKOR | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Potassium channel SKOR | | Authors: | Gao, X, Sun, T, Lu, Y, Jia, Y, Xu, X, Zhang, Y, Fu, P, Yang, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural changes in the conversion of an Arabidopsis outward-rectifying K + channel into an inward-rectifying channel.
Plant Commun., 5, 2024
|
|
8WUI
 
 | | SKOR D312N L271P double mutation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Potassium channel SKOR | | Authors: | Gao, X, Sun, T, Lu, Y, Jia, Y, Xu, X, Zhang, Y, Fu, P, Yang, G. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural changes in the conversion of an Arabidopsis outward-rectifying K + channel into an inward-rectifying channel.
Plant Commun., 5, 2024
|
|
8VQ3
 
 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-198. | | Descriptor: | (8R)-N-[(2S,3R)-3-(cyclohexylmethoxy)-1-(morpholin-4-yl)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-6-(1,3-thiazole-5-carbonyl)-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Expanding the ligandable proteome by paralog hopping with covalent probes.
Biorxiv, 2024
|
|
6J1M
 
 | |
8IOF
 
 | |
8ID1
 
 | |
8TWR
 
 | | Influenza A virus (A/Aichi/2/1968(H3N2) nucleoprotein mutant - 2-7 deleted, P283S, R416A | | Descriptor: | Nucleoprotein, SODIUM ION | | Authors: | Yoon, J, Zhang, Y.M, Grant, R.A, Shoulders, M.D. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The immune-evasive proline-283 substitution in influenza nucleoprotein increases aggregation propensity without altering the native structure.
Sci Adv, 10, 2024
|
|
8U2Y
 
 | | Solution structure of the PHD6 finger of MLL4 bound to TET3 | | Descriptor: | Histone-lysine N-methyltransferase 2D, Methylcytosine dioxygenase TET3, ZINC ION | | Authors: | Mohid, S.A, Zandian, M, Zhang, Y, Kutateladze, T.G. | | Deposit date: | 2023-09-06 | | Release date: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | MLL4 binds TET3.
Structure, 32, 2024
|
|
