7P0Z
 
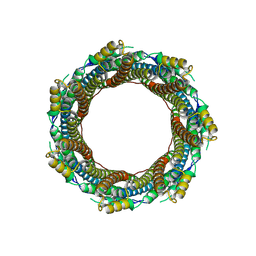 | | 2.43 A Mycobacterium marinum EspB. | | 分子名称: | ESX-1 secretion-associated protein EspB | | 著者 | Gijsbers, A, Zhang, Y, Vinciauskaite, V, Siroy, A, Ye, G, Tria, G, Mathew, A, Sanchez-Puig, N, Lopez-Iglesias, C, Peters, P.J, Ravelli, R.B.G. | | 登録日 | 2021-07-01 | | 公開日 | 2021-08-18 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (2.43 Å) | | 主引用文献 | Priming mycobacterial ESX-secreted protein B to form a channel-like structure.
Curr Res Struct Biol, 3, 2021
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5XKY
 
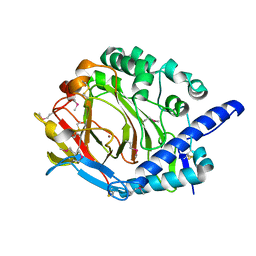 | | Crystal structure of DddY Se derivative | | 分子名称: | Uncharacterized protein, ZINC ION | | 著者 | Li, C.Y, Zhang, Y.Z. | | 登録日 | 2017-05-10 | | 公開日 | 2017-11-01 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | Mechanistic Insights into Dimethylsulfoniopropionate Lyase DddY, a New Member of the Cupin Superfamily.
J. Mol. Biol., 429, 2017
|
|
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | 分子名称: | S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
3VF9
 
 | |
6L4R
 
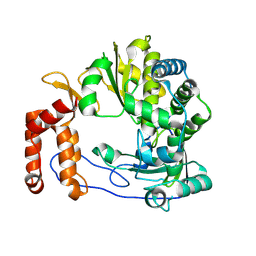 | | Crystal structure of Enterovirus D68 RdRp | | 分子名称: | RdRp | | 著者 | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | 登録日 | 2019-10-21 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.147 Å) | | 主引用文献 | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
3V8T
 
 | |
2RE9
 
 | | Crystal structure of TL1A at 2.1 A | | 分子名称: | GLYCEROL, MAGNESIUM ION, TNF superfamily ligand TL1A | | 著者 | Jin, T.C, Guo, F, Kim, S, Howard, A.J, Zhang, Y.Z. | | 登録日 | 2007-09-25 | | 公開日 | 2007-10-09 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | X-ray crystal structure of TNF ligand family member TL1A at 2.1 A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
3VF8
 
 | |
7SGV
 
 | | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder630 inhibitor | | 分子名称: | CHLORIDE ION, N-(naphthalen-1-yl)pyridine-3-carboxamide, Papain-like protease, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-10-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder630 inhibitor
To Be Published
|
|
7SGU
 
 | | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder608 inhibitor | | 分子名称: | 5-amino-N-(naphthalen-1-yl)pyridine-3-carboxamide, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-10-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder608 inhibitor
To Be Published
|
|
7SGW
 
 | | Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder630 inhibitor | | 分子名称: | CHLORIDE ION, N-(naphthalen-1-yl)pyridine-3-carboxamide, Non-structural protein 3, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-10-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder630 inhibitor
To Be Published
|
|
3V8W
 
 | |
4Q8K
 
 | | Crystal structure of polysaccharide lyase family 18 aly-SJ02 P-CATD | | 分子名称: | Alginase, CALCIUM ION, SULFATE ION | | 著者 | Dong, S, Li, C.Y, Zhang, Y.Z. | | 登録日 | 2014-04-28 | | 公開日 | 2014-09-17 | | 最終更新日 | 2015-11-25 | | 実験手法 | X-RAY DIFFRACTION (1.646 Å) | | 主引用文献 | Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18.
J.Biol.Chem., 289, 2014
|
|
4Q8L
 
 | | Crystal structure of polysacchride lyase family 18 aly-SJ02 r-CATD | | 分子名称: | Alginase, CALCIUM ION | | 著者 | Dong, S, Li, C.Y, Zhang, Y.Z. | | 登録日 | 2014-04-28 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18.
J.Biol.Chem., 289, 2014
|
|
5Y93
 
 | | Crystal Structure Analysis of the BRD4 | | 分子名称: | 1,2-ETHANEDIOL, 2-[[5-[(5-bromanyl-2-methoxy-phenyl)sulfonylamino]-3-methyl-1,2-benzoxazol-6-yl]oxy]-N-(2-morpholin-4-ylethyl)ethanamide, Bromodomain-containing protein 4, ... | | 著者 | Xu, Y, Zhang, Y, Song, M, Wang, C. | | 登録日 | 2017-08-22 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
1OQN
 
 | | Crystal structure of the phosphotyrosine binding domain (PTB) of mouse Disabled 1 (Dab1) | | 分子名称: | Alzheimer's disease amyloid A4 protein homolog, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Disabled homolog 1 | | 著者 | Yun, M, Keshvara, L, Park, C.-G, Zhang, Y.-M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.-W. | | 登録日 | 2003-03-10 | | 公開日 | 2003-08-05 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of the Dab homology domains of mouse disabled 1 and 2
J.Biol.Chem., 278, 2003
|
|
6AKT
 
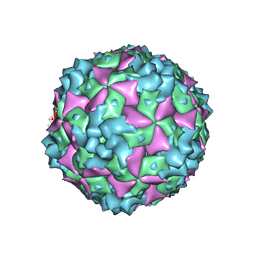 | | Cryo-EM structure of CVA10 A-particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | 登録日 | 2018-09-03 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKS
 
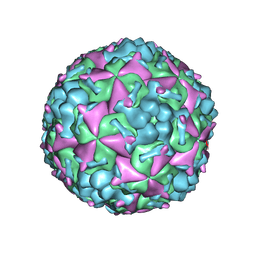 | | Cryo-EM structure of CVA10 mature virus | | 分子名称: | SPHINGOSINE, VP1, VP2, ... | | 著者 | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | 登録日 | 2018-09-03 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6ISM
 
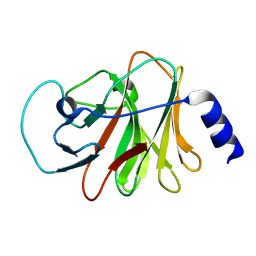 | | Crystal structure of intracellular B30.2 domain of BTN3A1 mutant | | 分子名称: | Butyrophilin subfamily 3 member A1 | | 著者 | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | 登録日 | 2018-11-16 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
5Y8W
 
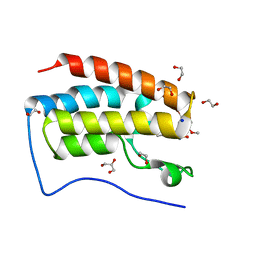 | | Crystal Structure Analysis of the BRD4 | | 分子名称: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(3-methyl-6-oxidanyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | 著者 | Xu, Y, Zhang, Y, Song, M, Wang, C. | | 登録日 | 2017-08-21 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y94
 
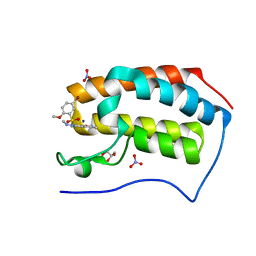 | | Crystal Structure Analysis of the BRD4 | | 分子名称: | 5-bromanyl-2-methoxy-N-[3-methyl-6-(methylamino)-1,2-benzoxazol-5-yl]benzenesulfonamide, Bromodomain-containing protein 4, GLYCEROL, ... | | 著者 | Xu, Y, Zhang, Y, Song, M, Wang, C. | | 登録日 | 2017-08-22 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
3CIV
 
 | |
8HNC
 
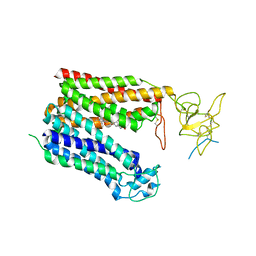 | | Cryo-EM structure of human OATP1B1 in complex with bilirubin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | 著者 | Shan, Z, Yang, X, Liu, H, Nan, J, Yuan, Y, Zhang, Y. | | 登録日 | 2022-12-07 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.73 Å) | | 主引用文献 | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
6AKU
 
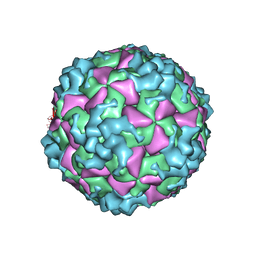 | | Cryo-EM structure of CVA10 empty particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | 登録日 | 2018-09-03 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
