8HUW
 
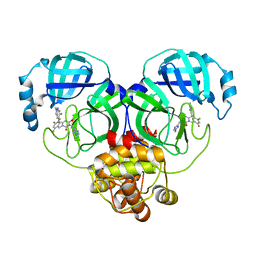 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with S217622 | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Wang, J, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
1RZQ
 
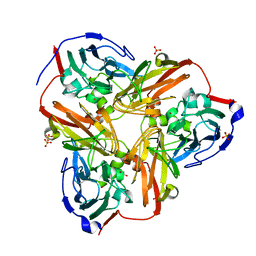 | | Crystal Structure of C-Terminal Despentapeptide Nitrite Reductase from Achromobacter Cycloclastes at pH5.0 | | 分子名称: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | 著者 | Li, H.T, Wang, C, Chang, T, Chang, W.C, Liu, M.Y, Le Gall, J, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | 登録日 | 2003-12-26 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | pH-profile crystal structure studies of C-terminal despentapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1RZP
 
 | | Crystal Structure of C-Terminal Despentapeptide Nitrite Reductase from Achromobacter Cycloclastes at pH6.2 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | 著者 | Li, H.T, Wang, C, Chang, T, Chang, W.C, Liu, M.Y, Le Gall, J, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | 登録日 | 2003-12-26 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | pH-profile crystal structure studies of C-terminal despentapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 316, 2004
|
|
5ITQ
 
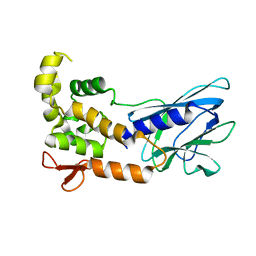 | | Crystal Structure of Human NEIL1, Free Protein | | 分子名称: | Endonuclease 8-like 1 | | 著者 | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | 登録日 | 2016-03-17 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8HVU
 
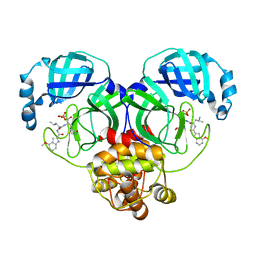 | |
8HVX
 
 | |
8HVV
 
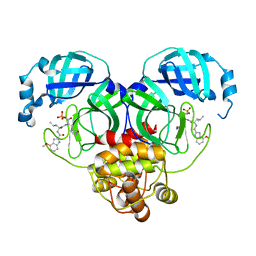 | |
8HVZ
 
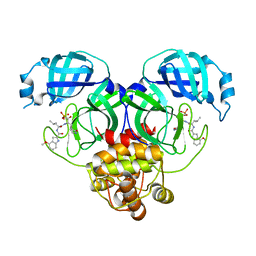 | |
8HVW
 
 | |
6J3Q
 
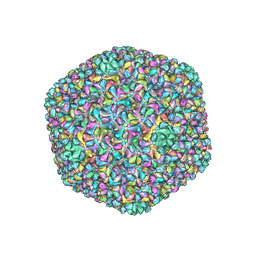 | | Capsid structure of a freshwater cyanophage Siphoviridae Mic1 | | 分子名称: | cement protein, major capsid protein | | 著者 | Jin, H, Jiang, Y.L, Yang, F, Zhang, J.T, Li, W.F, Zhou, K, Ju, J, Chen, Y, Zhou, C.Z. | | 登録日 | 2019-01-05 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Capsid Structure of a Freshwater Cyanophage Siphoviridae Mic1.
Structure, 27, 2019
|
|
3GGQ
 
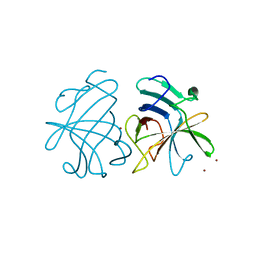 | | Dimerization of Hepatitis E Virus Capsid Protein E2s Domain is Essential for Virus-Host Interaction | | 分子名称: | BROMIDE ION, Capsid protein | | 著者 | Li, S.W, Tang, X.H, Seetharaman, J, Yang, C.Y, Gu, Y, Zhang, J, Du, H.L, Shih, J.W.K, Hew, C.L, Sivaraman, J, Xia, N.S. | | 登録日 | 2009-03-02 | | 公開日 | 2009-08-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Dimerization of hepatitis E virus capsid protein E2s domain is essential for virus-host interaction
Plos Pathog., 5, 2009
|
|
5VLZ
 
 | | Backbone model for phage Qbeta capsid | | 分子名称: | Capsid protein, Maturation protein A2 | | 著者 | Cui, Z, Zhang, J. | | 登録日 | 2017-04-26 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | 分子名称: | 3C-like proteinase | | 著者 | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | 登録日 | 2020-12-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
3IZI
 
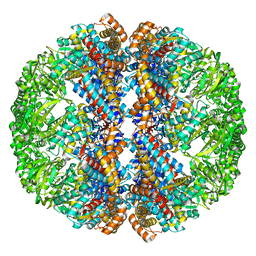 | | Mm-cpn rls with ATP | | 分子名称: | Chaperonin | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-29 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZH
 
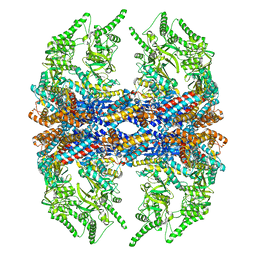 | | Mm-cpn D386A with ATP | | 分子名称: | Chaperonin | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-29 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
7YFG
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (major class in asymmetry) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | 著者 | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFH
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine, glutamate and (R)-PYD-106 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | 著者 | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3EMN
 
 | | The Crystal Structure of Mouse VDAC1 at 2.3 A resolution | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | 著者 | Ujwal, R, Cascio, D, Colletier, J.-P, Faham, S, Zhang, J, Toro, L, Ping, P, Abramson, J. | | 登録日 | 2008-09-24 | | 公開日 | 2008-12-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3LLP
 
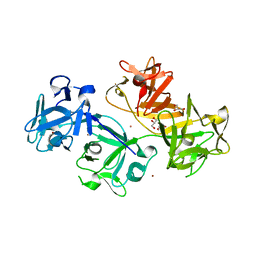 | | 1.8 Angstrom human fascin 1 crystal structure | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, Fascin, ... | | 著者 | Chen, L, Yang, S, Jakoncic, J, Zhang, J.J, Huang, X.-Y. | | 登録日 | 2010-01-29 | | 公開日 | 2010-04-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Migrastatin analogues target fascin to block tumour metastasis.
Nature, 464, 2010
|
|
4Q2W
 
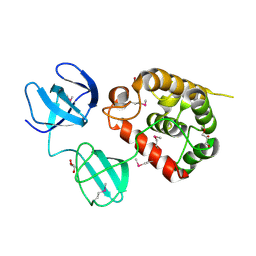 | | Crystal Structure of pneumococcal peptidoglycan hydrolase LytB | | 分子名称: | GLYCEROL, Putative endo-beta-N-acetylglucosaminidase | | 著者 | Bai, X.H, Chen, H.J, Jiang, Y.L, Wen, Z, Cheng, W, Li, Q, Zhang, J.R, Chen, Y, Zhou, C.Z. | | 登録日 | 2014-04-10 | | 公開日 | 2014-07-16 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure of pneumococcal peptidoglycan hydrolase LytB reveals insights into the bacterial cell wall remodeling and pathogenesis.
J.Biol.Chem., 289, 2014
|
|
5JIQ
 
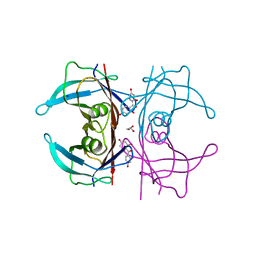 | | Crystal Structure of Human Transthyretin in Complex with 2,2',4,4'-tetrahydroxybenzophenone (BP2) | | 分子名称: | GLYCEROL, SODIUM ION, Transthyretin, ... | | 著者 | Begum, A, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | 登録日 | 2016-04-22 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
8UEJ
 
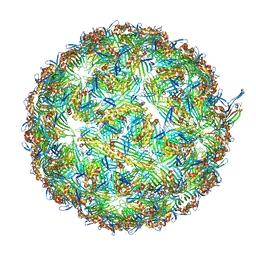 | | ssRNA phage PhiCb5 virion | | 分子名称: | CALCIUM ION, Coat protein, Maturation protein | | 著者 | Wang, Y, Zhang, J. | | 登録日 | 2023-10-01 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural mechanisms of Tad pilus assembly and its interaction with an RNA virus.
Sci Adv, 10, 2024
|
|
8HUX
 
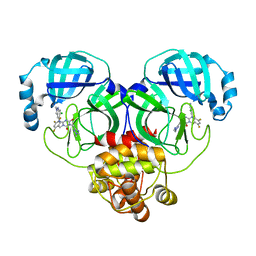 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with S217622 | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Li, W.W, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUR
 
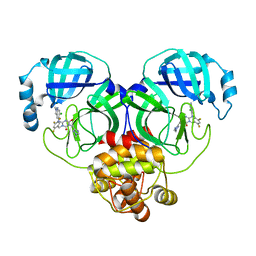 | | Crystal structure of SARS-Cov-2 main protease in complex with S217622 | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Zhou, X.L, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUS
 
 | | Crystal structure of SARS main protease in complex with S217622 | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Lin, C, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
