4O5Y
 
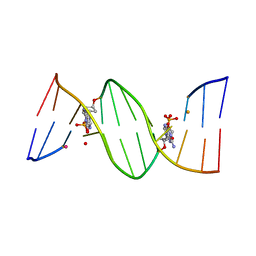 | | O6-carboxymethylguanine in DNA forms a sequence context dependent wobble base pair structure with thymine | | Descriptor: | BARIUM ION, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), POTASSIUM ION | | Authors: | Zhang, F, Tsunoda, M, Suzuki, K, Kikuchi, Y, Wilkinson, O, Millington, C.L, Margison, G.P, Williams, D.M, Takenaka, A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | O(6)-Carboxymethylguanine in DNA forms a sequence context-dependent wobble base-pair structure with thymine
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O5X
 
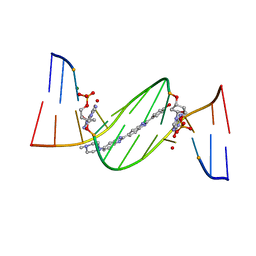 | | O6-carboxymethylguanine in DNA forms a sequence context dependent wobble base pair structure with thymine. | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Zhang, F, Tsunoda, M, Suzuki, K, Kikuchi, Y, Wilkinson, O, Millington, C.L, Margison, G.P, Williams, D.M, Takenaka, A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | O(6)-Carboxymethylguanine in DNA forms a sequence context-dependent wobble base-pair structure with thymine
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4YLY
 
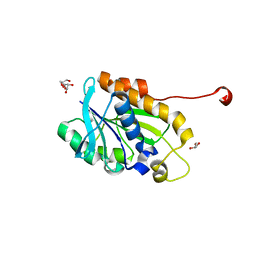 | | Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, staphylococcus aureus at 2.25 angstrom resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-03-06 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of Staphylococcus aureus peptidyl-tRNA hydrolase at a 2.25 angstrom resolution.
Acta Biochim.Biophys.Sin., 47, 2015
|
|
1KZX
 
 | | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) | | Descriptor: | INTESTINAL FATTY ACID-BINDING PROTEIN (T54) | | Authors: | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) that is associated with altered lipid metabolism
Biochemistry, 42, 2003
|
|
1KZW
 
 | | Solution structure of Human Intestinal Fatty acid binding protein | | Descriptor: | INTESTINAL FATTY ACID-BINDING PROTEIN (A54) | | Authors: | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) that is associated with altered lipid metabolism
Biochemistry, 42, 2003
|
|
1AX8
 
 | | Human obesity protein, leptin | | Descriptor: | OBESITY PROTEIN | | Authors: | Zhang, F, Beals, J.M, Briggs, S.L, Clawson, D.K, Wery, J.-P, Schevitz, R.W. | | Deposit date: | 1997-10-31 | | Release date: | 1998-11-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the obese protein leptin-E100.
Nature, 387, 1997
|
|
6RP8
 
 | |
1PYE
 
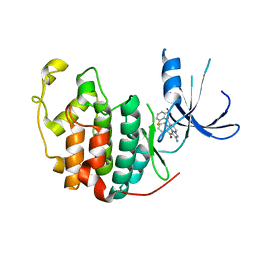 | | Crystal structure of CDK2 with inhibitor | | Descriptor: | Cell division protein kinase 2, [2-AMINO-6-(2,6-DIFLUORO-BENZOYL)-IMIDAZO[1,2-A]PYRIDIN-3-YL]-PHENYL-METHANONE | | Authors: | Zhang, F, Hamdouchi, C. | | Deposit date: | 2003-07-08 | | Release date: | 2004-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The discovery of a new structural class of cyclin-dependent kinase inhibitors, aminoimidazo[1,2-a]pyridines.
MOL.CANCER THER., 3, 2004
|
|
9JA8
 
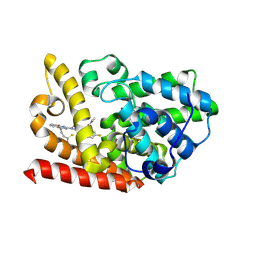 | | Crystal structure of human phosphodiesterase 10A in complex with N-(2-amino-2-thioxoethyl)-2-(3-(3-(dimethylcarbamoyl)-6-fluoroimidazo[1,2-a]pyridin-2-yl)azetidin-1-yl)quinoline-4-carboxamide | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, ... | | Authors: | Zhang, F.C, Huang, Y.Y, Luo, H.B, Guo, L. | | Deposit date: | 2024-08-24 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhalable Carbonyl Sulfide Donor-Hybridized Selective Phosphodiesterase 10A Inhibitor for Treating Idiopathic Pulmonary Fibrosis by Inhibiting Tumor Growth Factor-beta Signaling and Activating the cAMP/Protein Kinase A/cAMP Response Element-Binding Protein (CREB)/p53 Axis.
Acs Pharmacol Transl Sci, 8, 2025
|
|
1PY5
 
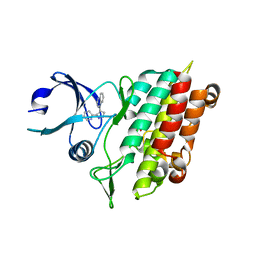 | | Crystal Structure of TGF-beta receptor I kinase with inhibitor | | Descriptor: | 4-(3-PYRIDIN-2-YL-1H-PYRAZOL-4-YL)QUINOLINE, SULFATE ION, TGF-beta receptor type I | | Authors: | Zhang, F, Sawyer, J.S. | | Deposit date: | 2003-07-08 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and activity of new aryl- and heteroaryl-substituted 5,6-dihydro-4H-pyrrolo[1,2-b]pyrazole inhibitors of the transforming growth factor-beta type I receptor kinase domain.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
5GXT
 
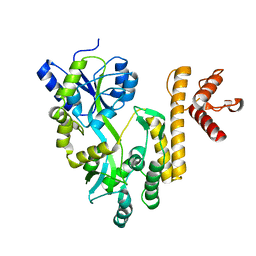 | | Crystal structure of PigG | | Descriptor: | MAGNESIUM ION, Maltose-binding periplasmic protein,PigG | | Authors: | Zhang, F, Ran, T, Xu, D, Wang, W. | | Deposit date: | 2016-09-20 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Crystal structure of MBP-PigG fusion protein and the essential function of PigG in the prodigiosin biosynthetic pathway in Serratia marcescens FS14.
Int. J. Biol. Macromol., 99, 2017
|
|
5GXV
 
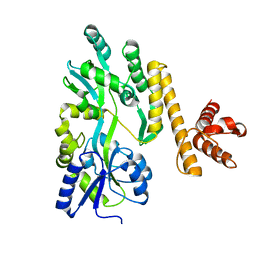 | | Crystal structure of PigG | | Descriptor: | MAGNESIUM ION, Maltose-binding periplasmic protein,PigG | | Authors: | Zhang, F, Ran, T, Xu, D, Wang, W. | | Deposit date: | 2016-09-20 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of MBP-PigG fusion protein and the essential function of PigG in the prodigiosin biosynthetic pathway in Serratia marcescens FS14.
Int. J. Biol. Macromol., 99, 2017
|
|
1RW8
 
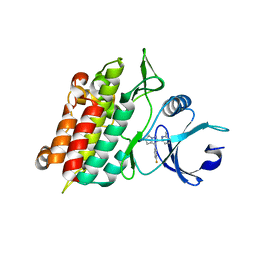 | | Crystal Structure of TGF-beta receptor I kinase with ATP site inhibitor | | Descriptor: | 3-(4-FLUOROPHENYL)-2-(6-METHYLPYRIDIN-2-YL)-5,6-DIHYDRO-4H-PYRROLO[1,2-B]PYRAZOLE, TGF-beta receptor type I | | Authors: | Zhang, F, Sawyer, J.S. | | Deposit date: | 2003-12-16 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and activity of new aryl- and heteroaryl-substituted 5,6-dihydro-4H-pyrrolo[1,2-b]pyrazole inhibitors of the transforming growth factor-beta type I receptor kinase domain.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
5CN1
 
 | |
8ZXD
 
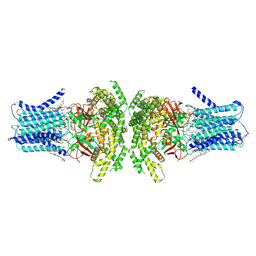 | | the Planar Cell Polarity Core Protein Vangl1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, Vang-like protein 1 | | Authors: | Zhang, F, Chen, S. | | Deposit date: | 2024-06-14 | | Release date: | 2024-11-27 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure and oligomerization of the human planar cell polarity core protein Vangl1.
Nat Commun, 16, 2025
|
|
5HS5
 
 | |
5J08
 
 | |
5CMW
 
 | | Crystal structure of yeast Ent5 N-terminal domain-soaked in KI | | Descriptor: | Epsin-5, GLYCEROL, IODIDE ION | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insight into the N-terminal domain of the clathrin adaptor Ent5 from Saccharomyces cerevisiae
Biochem.Biophys.Res.Commun., 477, 2016
|
|
5CMY
 
 | | Crystal structure of yeast Ent5 N-terminal domain-native | | Descriptor: | Epsin-5, GLYCEROL | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and functional insight into the N-terminal domain of the clathrin adaptor Ent5 from Saccharomyces cerevisiae
Biochem.Biophys.Res.Commun., 477, 2016
|
|
3IFB
 
 | | NMR STUDY OF HUMAN INTESTINAL FATTY ACID BINDING PROTEIN | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN | | Authors: | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | Deposit date: | 1998-10-16 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human intestinal fatty acid binding protein: implications for ligand entry and exit.
J.Biomol.NMR, 9, 1997
|
|
4IJ0
 
 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*CP*GP*CP*G)-3'), STRONTIUM ION | | Authors: | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
5CN2
 
 | |
2K23
 
 | |
4ITD
 
 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*(C6G)P*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | Deposit date: | 2013-01-18 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
4O5W
 
 | | O6-carboxymethylguanine in DNA forms a sequence context dependent wobble base pair structure with thymine | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Zhang, F, Tsunoda, M, Suzuki, K, Kikuchi, Y, Wilkinson, O, Millington, C.L, Margison, G.P, Williams, D.M, Takenaka, A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | O(6)-Carboxymethylguanine in DNA forms a sequence context-dependent wobble base-pair structure with thymine
Acta Crystallogr.,Sect.D, 70, 2014
|
|
