6LAD
 
 | |
5IZU
 
 | |
6LAF
 
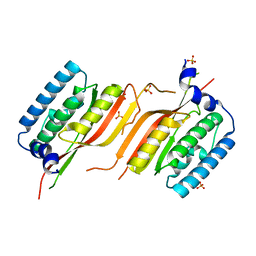 | | Crystal structure of the core domain of Amuc_1100 from Akkermansia muciniphila | | Descriptor: | Amuc_1100, SULFATE ION | | Authors: | Wang, J, Xiang, R, Zhang, M, Wang, M. | | Deposit date: | 2019-11-12 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | The variable oligomeric state of Amuc_1100 from Akkermansia muciniphila.
J.Struct.Biol., 212, 2020
|
|
4LH9
 
 | |
5JXB
 
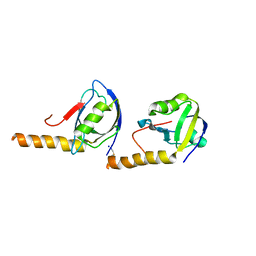 | | PSD-95 extended PDZ3 in complex with SynGAP PBM | | Descriptor: | Disks large homolog 4,SynGAP | | Authors: | Shang, Y, Zhang, M. | | Deposit date: | 2016-05-13 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity.
Cell, 166, 2016
|
|
2P91
 
 | | Crystal structure of Enoyl-[acyl-carrier-protein] reductase (NADH) from Aquifex aeolicus VF5 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Chen, L, Li, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Enoyl-[acyl-carrier-protein] reductase (NADH) from Aquifex aeolicus VF5
To be Published
|
|
5JXC
 
 | | SynGAP Coiled-coil trimer | | Descriptor: | Ras/Rap GTPase-activating protein SynGAP | | Authors: | Shang, Y, Zhang, M. | | Deposit date: | 2016-05-13 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity.
Cell, 166, 2016
|
|
6XOU
 
 | |
6XOV
 
 | |
6XOS
 
 | |
6XOT
 
 | |
2O4X
 
 | | Crystal structure of human P100 tudor domain | | Descriptor: | Staphylococcal nuclease domain-containing protein 1 | | Authors: | Shaw, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Rao, Z, Wang, B.C, Liu, Z.J. | | Deposit date: | 2006-12-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human P100 tudor domain
To be Published
|
|
2P5U
 
 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Dillard, B, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
5IH5
 
 | | Human Casein Kinase 1 isoform delta (kinase domain) in complex with Epiblastin A | | Descriptor: | 6-(3-chlorophenyl)pteridine-2,4,7-triamine, Casein kinase I isoform delta, S,R MESO-TARTARIC ACID, ... | | Authors: | Ursu, A, Illich, D.J, Takemoto, Y, Porfetye, A.T, Zhang, M, Brockmeyer, A, Janning, P, Watanabe, N, Osada, H, Vetter, I.R, Ziegler, S, Schoeler, H.R, Waldmann, H. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epiblastin A Induces Reprogramming of Epiblast Stem Cells Into Embryonic Stem Cells by Inhibition of Casein Kinase 1.
Cell Chem Biol, 23, 2016
|
|
8IH4
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN2A2 Mutant | | Descriptor: | Butyrophilin subfamily 2 member A2 | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2023-02-22 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
2PG0
 
 | | Crystal structure of acyl-CoA dehydrogenase from Geobacillus kaustophilus | | Descriptor: | Acyl-CoA dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Chen, L, Chen, L.-Q, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Li, Y, Fu, Z.-Q, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of acyl-CoA dehydrogenase from G. kaustophilus
To be Published
|
|
8IW4
 
 | | Cryo-EM structure of the SPE-bound mTAAR9-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW7
 
 | | Cryo-EM structure of the PEA-bound mTAAR9-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW9
 
 | | Cryo-EM structure of the CAD-bound mTAAR9-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8ITF
 
 | | Cryo-EM structure of the DMCHA-bound mTAAR9-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-22 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IWE
 
 | | Cryo-EM structure of the SPE-mTAAR9 complex | | Descriptor: | SPERMIDINE, Trace amine-associated receptor 9 | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW1
 
 | | Cryo-EM structure of the PEA-bound mTAAR9-Golf complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(olf) subunit alpha, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IWM
 
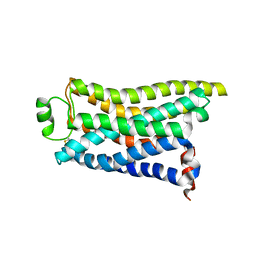 | | Cryo-EM structure of the PEA-bound mTAAR9 complex | | Descriptor: | 2-PHENYLETHYLAMINE, Trace amine-associated receptor 9 | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
4JHA
 
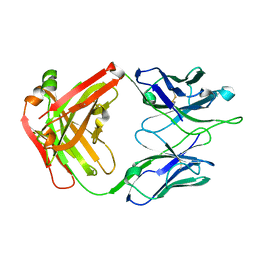 | | Crystal Structure of RSV-Neutralizing Human Antibody D25 | | Descriptor: | D25 antigen-binding fragment heavy chain, D25 light chain | | Authors: | Mclellan, J.S, Chen, M, Leung, S, Graepel, K.W, Du, X, Yang, Y, Zhou, T, Baxa, U, Yasuda, E, Beaumont, T, Kumar, A, Modjarrad, K, Zheng, Z, Zhao, M, Xia, N, Kwong, P.D, Graham, B.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody.
Science, 340, 2013
|
|
8F4F
 
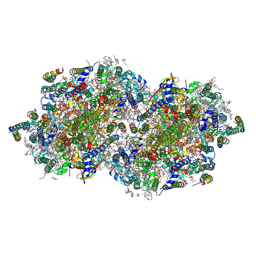 | | RT XFEL structure of Photosystem II 500 microseconds after the third illumination at 2.03 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
