3L82
 
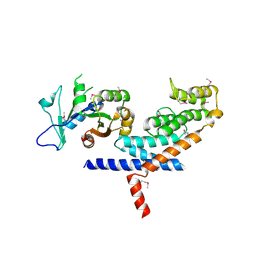 | | X-ray Crystal structure of TRF1 and Fbx4 complex | | Descriptor: | F-box only protein 4, Telomeric repeat-binding factor 1 | | Authors: | Zeng, Z.X, Wang, W, Yang, Y.T, Chen, Y, Yang, X.M, Diehl, J.A, Liu, X.D, Lei, M. | | Deposit date: | 2009-12-29 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Selective Ubiquitination of TRF1 by SCF(Fbx4)
Dev.Cell, 18, 2010
|
|
1GIK
 
 | | POKEWEED ANTIVIRAL PROTEIN FROM SEEDS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIVIRAL PROTEIN S | | Authors: | Zeng, Z.H, He, X.L, Li, H.M, Hu, Z, Wang, D.C. | | Deposit date: | 2001-02-07 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of pokeweed antiviral protein with well-defined sugars from seeds at 1.8 angstrom resolution
J.Struct.Biol., 141, 2003
|
|
1CD1
 
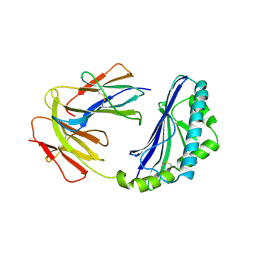 | |
3U50
 
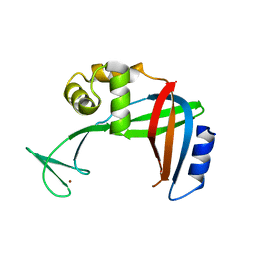 | | Crystal Structure of the Tetrahymena telomerase processivity factor Teb1 OB-C | | Descriptor: | Telomerase-associated protein 82, ZINC ION | | Authors: | Zeng, Z, Huang, J, Yang, Y, Lei, M. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U58
 
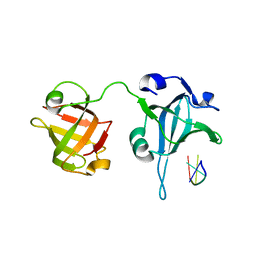 | | Crystal Structure of the Tetrahymena telomerase processivity factor Teb1 AB | | Descriptor: | DNA (5'-D(*GP*GP*GP*T)-3'), Tetrahymena Teb1 AB | | Authors: | Zeng, Z, Huang, J, Yang, Y, Lei, M. | | Deposit date: | 2011-10-11 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U4V
 
 | |
3U4Z
 
 | |
7Y6C
 
 | | Crystal structure of the EscE/EsaG/EsaH complex | | Descriptor: | EscE/YscE/SsaE family type III secretion system needle protein co-chaperone, EscG/YscG/SsaH family type III secretion system needle protein co-chaperone | | Authors: | Zeng, Z.X, Xiao, S.B, Liao, L.J, Zhou, Y.Z. | | Deposit date: | 2022-06-18 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Secreted in a Type III Secretion System-Dependent Manner, EsaH and EscE Are the Cochaperones of the T3SS Needle Protein EsaG of Edwardsiella piscicida.
Mbio, 13, 2022
|
|
7Y6B
 
 | | Crystal structure of the EscE/EsaH complex | | Descriptor: | EscE/YscE/SsaE family type III secretion system needle protein co-chaperone, EscG/YscG/SsaH family type III secretion system needle protein co-chaperone | | Authors: | Zeng, Z.X, Xiao, S.B, Liao, L.J, Zhou, Y.Z. | | Deposit date: | 2022-06-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Secreted in a Type III Secretion System-Dependent Manner, EsaH and EscE Are the Cochaperones of the T3SS Needle Protein EsaG of Edwardsiella piscicida.
Mbio, 13, 2022
|
|
5YM8
 
 | |
5YM6
 
 | |
8IBI
 
 | | Inactive mutant of CtPL-H210S/F214I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
8IBJ
 
 | | Inactive mutant of CtPL-H210S/F214I/N181A/F235L | | Descriptor: | PET hydrolase | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
8IAN
 
 | | Crystal structure of CtPL-H210S/F214I mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PET hydrolase | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-08 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
6K5M
 
 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa (Rice) | | Descriptor: | Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-05-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6KHO
 
 | | Crystal structure of Oryza sativa TDC with PLP | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Crystal structure ofOryza sativaTDC reveals the substrate specificity for TDC-mediated melatonin biosynthesis.
J Adv Res, 24, 2020
|
|
6KHN
 
 | | Crystal structure of Oryza sativa TDC with PLP and SEROTONIN | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crystal structure ofOryza sativaTDC reveals the substrate specificity for TDC-mediated melatonin biosynthesis.
J Adv Res, 24, 2020
|
|
1SN4
 
 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M4 | | Descriptor: | ACETATE ION, PROTEIN (NEUROTOXIN BMK M4) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1SNB
 
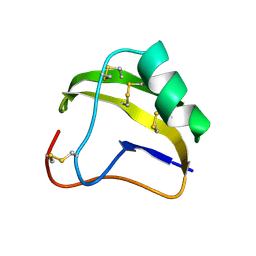 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M8 | | Descriptor: | NEUROTOXIN BMK M8 | | Authors: | Wang, D.C, Zeng, Z.H, Li, H.M. | | Deposit date: | 1997-03-12 | | Release date: | 1997-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an acidic neurotoxin from scorpion Buthus martensii Karsch at 1.85 A resolution.
J.Mol.Biol., 261, 1996
|
|
1SN1
 
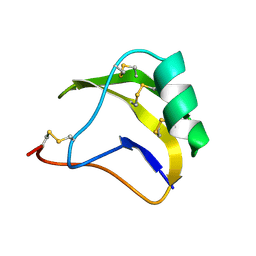 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M1 | | Descriptor: | PROTEIN (NEUROTOXIN BMK M1) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
6K5S
 
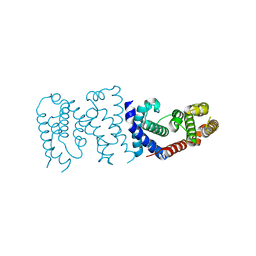 | |
6K5U
 
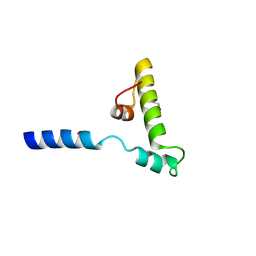 | |
2RFH
 
 | |
8ZR5
 
 | | Cryo-EM Structure of GPR119-Gs-Firuglipel complex | | Descriptor: | 4-[5-[(1~{R})-1-(4-cyclopropylcarbonylphenoxy)propyl]-1,2,4-oxadiazol-3-yl]-2-fluoranyl-~{N}-[(2~{R})-1-oxidanylpropan-2-yl]benzamide, Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wong, T.S, Zeng, Z.C, Xiong, T.T, Gan, S.Y, Qiu, C, Du, Y. | | Deposit date: | 2024-06-04 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM structure of GPR119-Gs complex
To Be Published
|
|
8ZRK
 
 | | Cryo-EM structure of GPR119-Gs Complex with small molecule agonist GSK-1292263 | | Descriptor: | GSK-1292263, Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wong, T.S, Xiong, T.T, Zeng, Z.C, Gan, S.Y, Qiu, C, Du, Y. | | Deposit date: | 2024-06-04 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-EM structure of GPR119-Gs complex
To be published
|
|
