5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5CMS
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, SULFATE ION | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5BOE
 
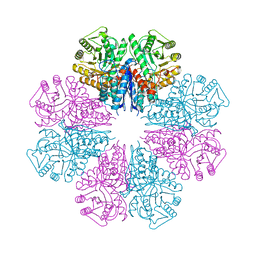 | | Crystal structure of Staphylococcus aureus enolase in complex with PEP | | Descriptor: | Enolase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wang, C.L, Wu, Y.F, Han, L, Wu, M.H, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5BOF
 
 | | Crystal Structure of Staphylococcus aureus Enolase | | Descriptor: | Enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Wu, Y.F, Wang, C.L, Wu, M.H, Han, L, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2GP3
 
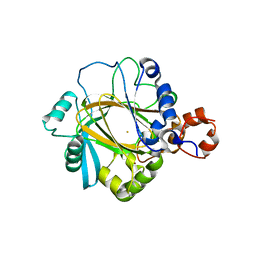 | | Crystal structure of the catalytic core domain of jmjd2a | | Descriptor: | FE (II) ION, Jumonji domain-containing protein 2A, ZINC ION | | Authors: | Chen, Z, Zang, J, Whetstine, J, Hong, X, Davrazou, F, Kutateladze, T.G, Simpson, M, Dai, S, Hagman, J, Shi, Y, Zhang, G. | | Deposit date: | 2006-04-16 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into histone demethylation by JMJD2 family members
Cell(Cambridge,Mass.), 125, 2006
|
|
3UQH
 
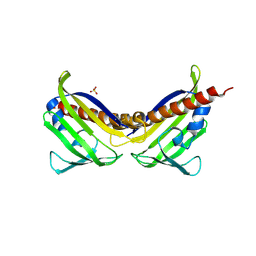 | | Crystal structure of aba receptor pyl10 (apo) | | Descriptor: | Abscisic acid receptor PYL10, SULFATE ION | | Authors: | Sun, D.M, Wang, H.P, Wu, M.H, Zang, J.Y, Tian, C.L. | | Deposit date: | 2011-11-20 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Aba Receptor Pyl10
To be Published
|
|
2GP5
 
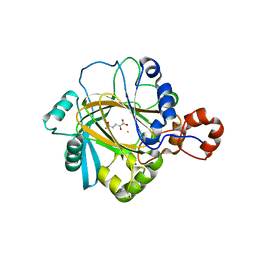 | | Crystal structure of catalytic core domain of jmjd2A complexed with alpha-Ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, Jumonji domain-containing protein 2A, ... | | Authors: | Chen, Z, Zang, J, Whetstine, J, Hong, X, Davrazou, F, Kutateladze, T.G, Simpson, M, Dai, S, Hagman, J, Shi, Y, Zhang, G. | | Deposit date: | 2006-04-16 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into histone demethylation by JMJD2 family members
Cell(Cambridge,Mass.), 125, 2006
|
|
5GOU
 
 | | Structure of a 16-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | Descriptor: | Ferritin heavy chain | | Authors: | Zhang, S, Zang, J, Wang, W, Wang, H, Zhao, G. | | Deposit date: | 2016-07-29 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Conversion of the Native 24-mer Ferritin Nanocage into Its Non-Native 16-mer Analogue by Insertion of Extra Amino Acid Residues.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5GN8
 
 | | Structure of a 48-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | Descriptor: | CALCIUM ION, Ferritin heavy chain | | Authors: | Zhang, S, Zang, J, Zhao, G, Mikami, B. | | Deposit date: | 2016-07-19 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | "Silent" Amino Acid Residues at Key Subunit Interfaces Regulate the Geometry of Protein Nanocages
ACS Nano, 10, 2016
|
|
2P5B
 
 | | The complex structure of JMJD2A and trimethylated H3K36 peptide | | Descriptor: | FE (II) ION, Histone H3, JmjC domain-containing histone demethylation protein 3A, ... | | Authors: | Zhang, G, Chen, Z, Zang, J, Hong, X, Shi, Y. | | Deposit date: | 2007-03-14 | | Release date: | 2007-06-12 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of the recognition of a methylated histone tail by JMJD2A.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4Y3O
 
 | | Crystal structure of Ribosomal oxygenase NO66 in complex with substrate Rpl8 peptide and Ni(II) and cofactor N-oxalyglycine | | Descriptor: | ACETATE ION, Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, GLYCEROL, ... | | Authors: | Wang, C, Zhang, Q, Zang, J. | | Deposit date: | 2015-02-10 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3R93
 
 | | Crystal structure of the chromo domain of M-phase phosphoprotein 8 bound to H3K9Me3 peptide | | Descriptor: | H3K9Me3 peptide, M-phase phosphoprotein 8, UNKNOWN ATOM OR ION | | Authors: | Li, J, Li, Z, Ruan, J, Xu, C, Tong, Y, Pan, P.W, Tempel, W, Crombet, L, Min, J, Zang, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Plos One, 6, 2011
|
|
3RY7
 
 | | Crystal Structure of Sa239 | | Descriptor: | GLYCEROL, Ribokinase | | Authors: | Li, J, Wu, M, Wang, L, Zang, J. | | Deposit date: | 2011-05-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Sa239 reveals the structural basis for the activation of ribokinase by monovalent cations.
J.Struct.Biol., 177, 2012
|
|
3P12
 
 | | Crystal Structure of D-ribose Pyranase Sa240 | | Descriptor: | D-ribose pyranase, GLYCEROL | | Authors: | Wu, M, Wang, L, Zang, J. | | Deposit date: | 2010-09-29 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Sa240: A ribose pyranase homolog with partial active site from Staphylococcus aureus
J.Struct.Biol., 174, 2011
|
|
3P13
 
 | |
4Y4R
 
 | | Crystal structure of ribosomal oxygenase NO66 dimer mutant | | Descriptor: | ACETATE ION, Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, NICKEL (II) ION | | Authors: | Wang, C, Hang, T, Zang, J. | | Deposit date: | 2015-02-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4E4H
 
 | | Crystal structure of Histone Demethylase NO66 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Wu, M, Tao, Y, Zang, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | tructural Basis for Tetramerization-dependent Gene Repression by Histone Demethylase NO66
To be Published
|
|
4GO1
 
 | | Crystal Structure of full length transcription repressor LsrR from E. coli. | | Descriptor: | GLYCEROL, Transcriptional regulator LsrR | | Authors: | Wu, M, Tao, Y, Liu, X, Zang, J. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Phosphorylated Autoinducer-2 Modulation of the Oligomerization State of the Global Transcription Regulator LsrR from Escherichia coli
J.Biol.Chem., 288, 2013
|
|
4GB0
 
 | |
4GAZ
 
 | | Crystal Structure of a Jumonji Domain-containing Protein JMJD5 | | Descriptor: | Lysine-specific demethylase 8, N-OXALYLGLYCINE, NICKEL (II) ION | | Authors: | Wang, H, Zhou, X, Zhang, X, Tao, Y, Chen, N, Zang, J. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of a Jumonji Domain-containing Protein JMJD5
To be Published
|
|
1WT9
 
 | | crystal structure of Aa-X-bp-I, a snake venom protein with the activity of binding to coagulation factor X from Agkistrodon acutus | | Descriptor: | CALCIUM ION, agkisacutacin A chain, anticoagulant protein-B | | Authors: | Zhu, Z, Liu, S, Mo, X, Yu, X, Liang, Z, Zang, J, Zhao, W, Teng, M, Niu, L. | | Deposit date: | 2004-11-18 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterizations and Crystal structures of two snake venom proteins with the activity of binding coagulation factor X from Agkistrodon acutus
To be Published
|
|
7WQL
 
 | | Bovin Beta-lactoglobulin binding with zinc ions | | Descriptor: | Beta-lactoglobulin, ZINC ION | | Authors: | Li, T, Ma, J, Zang, J, Zhao, G, Zhang, T. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Zinc binding strength of proteins dominants zinc uptake in Caco-2 cells.
Rsc Adv, 12, 2022
|
|
3R6P
 
 | | Crystal structure of abscisic acid-bound PYL10 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL10 | | Authors: | Sun, D.M, Wu, M.H, Wang, H.P, Zang, J.Y, Tian, C.L. | | Deposit date: | 2011-03-22 | | Release date: | 2011-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of abscisic acid-bound PYL10
To be Published
|
|
1Y17
 
 | | crystal structure of Aa-X-bp-II, a snake venom protein with the activity of binding to coagulation factor X from Agkistrodon acutus | | Descriptor: | CALCIUM ION, anticoagulant protein A, anticoagulant protein-B | | Authors: | Zhu, Z, Liu, S, Mo, X, Yu, X, Liang, Z, Zang, J, Zhao, W, Teng, M, Niu, L. | | Deposit date: | 2004-11-17 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterizations and Crystal structures of two snake venom proteins with the activity of binding coagulation factor X from Agkistrodon acutus
To be Published
|
|
3LD8
 
 | | Structure of JMJD6 and Fab Fragments | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, FE (III) ION, GLYCEROL, ... | | Authors: | Hong, X, Zang, J, White, J, Kappler, J.W, Wang, C, Zhang, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-08-04 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction of JMJD6 with single-stranded RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
