2RIM
 
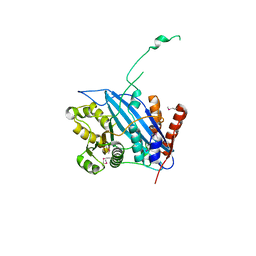 | | Crystal structure of Rtt109 | | Descriptor: | Regulator of Ty1 transposition protein 109 | | Authors: | Yuan, Y.A. | | Deposit date: | 2007-10-12 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into histone h3 lysine 56 acetylation by rtt109
Structure, 16, 2008
|
|
3VZ3
 
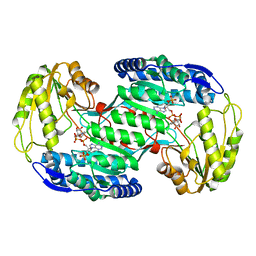 | | Structural insights into substrate and cofactor selection by sp2771 | | Descriptor: | 4-oxobutanoic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase | | Authors: | Yuan, Y.A, Yuan, Z, Yin, B, Wei, D. | | Deposit date: | 2012-10-09 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis for cofactor and substrate selection by cyanobacterium succinic semialdehyde dehydrogenase
J.Struct.Biol., 182, 2013
|
|
3VZ0
 
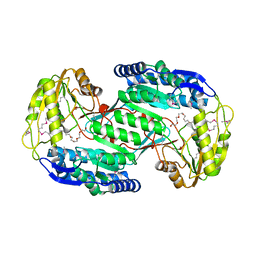 | | Structural insights into cofactor and substrate selection by Gox0499 | | Descriptor: | NONAETHYLENE GLYCOL, Putative NAD-dependent aldehyde dehydrogenase | | Authors: | Yuan, Y.A, Yuan, Z, Yin, B, Wei, D. | | Deposit date: | 2012-10-09 | | Release date: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for cofactor and substrate selection by cyanobacterium succinic semialdehyde dehydrogenase
J.Struct.Biol., 182, 2013
|
|
3VZ1
 
 | |
3WA8
 
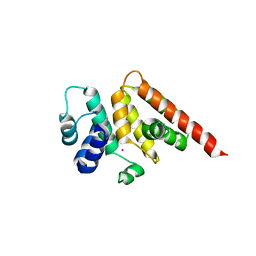 | | Crystal structure of M. ruber CasB | | Descriptor: | CRISPR-associated protein, Cse2 family, MERCURY (II) ION | | Authors: | Yuan, Y.A, Yuan, Z. | | Deposit date: | 2013-04-28 | | Release date: | 2014-04-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into crRNA G-rich sequence binding and R-loop formation facilitated by Meiothermus ruber CasB
To be Published
|
|
3VZ2
 
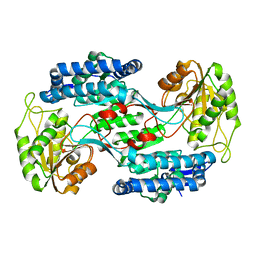 | |
2F8S
 
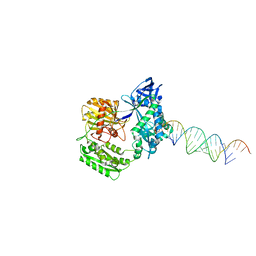 | | Crystal structure of Aa-Ago with externally-bound siRNA | | Descriptor: | 5'-R(P*AP*GP*AP*CP*AP*GP*CP*AP*UP*AP*UP*AP*UP*GP*CP*UP*GP*UP*CP*UP*UP*U)-3', Argonaute protein | | Authors: | Yuan, Y.R, Chen, H.Y, Patel, D.J. | | Deposit date: | 2005-12-03 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Potential Protein-RNA Recognition Event along the RISC-Loading Pathway from the Structure of A. aeolicus Argonaute with Externally Bound siRNA.
Structure, 14, 2006
|
|
6L8Q
 
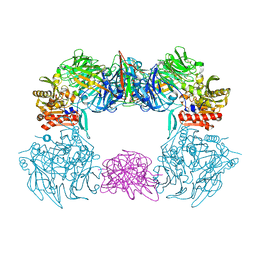 | | Complex structure of bat CD26 and MERS-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Yuan, Y. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Basis of Binding between Middle East Respiratory Syndrome Coronavirus and CD26 from Seven Bat Species.
J.Virol., 94, 2020
|
|
6OQK
 
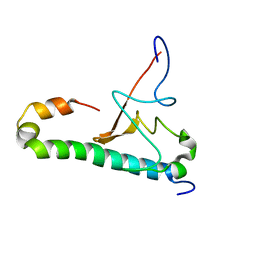 | |
6OQ9
 
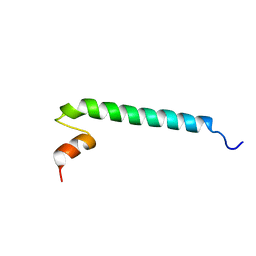 | |
6OKY
 
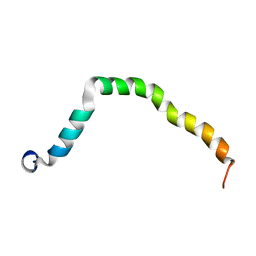 | | Solution structure of truncated peptide from PAMap53 | | Descriptor: | Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Yuan, Y, Castellino, F.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structural model of the complex of the binding regions of human plasminogen with its M-protein receptor from Streptococcus pyogenes.
J.Struct.Biol., 208, 2019
|
|
6OKX
 
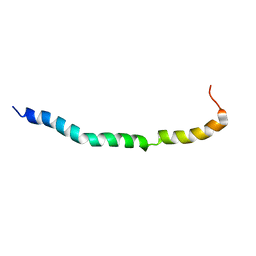 | | Solution structure of VEK50RH1/AA | | Descriptor: | Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Yuan, Y, Castellino, F.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structural model of the complex of the binding regions of human plasminogen with its M-protein receptor from Streptococcus pyogenes.
J.Struct.Biol., 208, 2019
|
|
6OQJ
 
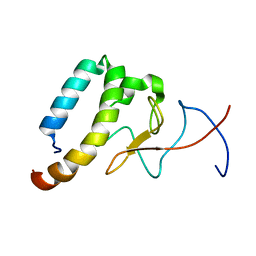 | |
6OKW
 
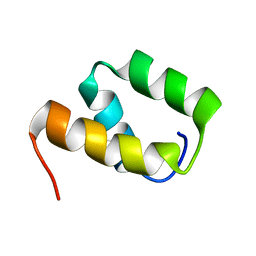 | | Solution structure of VEK50 | | Descriptor: | Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Yuan, Y, Castellino, F.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structural model of the complex of the binding regions of human plasminogen with its M-protein receptor from Streptococcus pyogenes.
J.Struct.Biol., 208, 2019
|
|
5B1Z
 
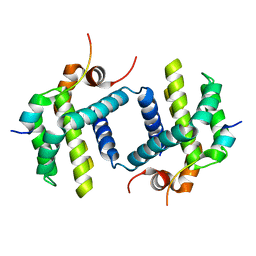 | |
5B4M
 
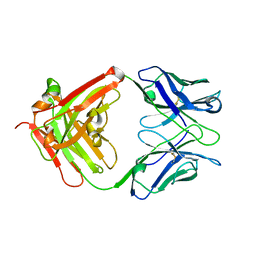 | |
1MV5
 
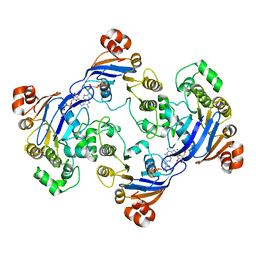 | | Crystal structure of LmrA ATP-binding domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yuan, Y, Chen, H, Patel, D. | | Deposit date: | 2002-09-24 | | Release date: | 2003-12-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of LmrA ATP-binding domain reveals the two-site alternating mechanism at molecular level
To be Published
|
|
1OSW
 
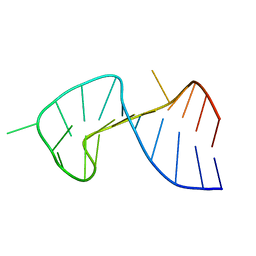 | | The Stem of SL1 RNA in HIV-1: Structure and Nucleocapsid Protein Binding for a 1X3 Internal Loop | | Descriptor: | 5'-R(*GP*GP*AP*GP*GP*CP*GP*CP*UP*AP*CP*GP*GP*CP*GP*AP*GP*GP*CP*UP*CP*CP*A)-3' | | Authors: | Yuan, Y, Kerwood, D.J, Paoletti, A.C, Shubsda, M.F, Borer, P.N. | | Deposit date: | 2003-03-20 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Stem of SL1 RNA in HIV-1: Structure and Nucleocapsid Protein Binding for a 1X3 Internal Loop
Biochemistry, 42, 2003
|
|
2ZFN
 
 | |
3D3H
 
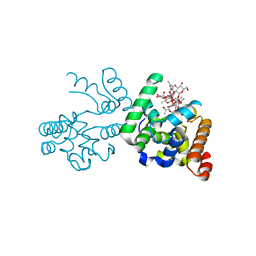 | | Crystal structure of a complex of the peptidoglycan glycosyltransferase domain from Aquifex aeolicus and neryl moenomycin A | | Descriptor: | (2R)-3-{[(S)-{[(2R,3R,4R,5S,6S)-3-{[(2S,3R,4R,5S,6R)-3-(acetylamino)-5-{[(2S,3R,4R,5S,6R)-3-(acetylamino)-5-{[(2R,3R,4S,5R,6S)-6-carbamoyl-3,4,5-trihydroxytetrahydro-2H-pyran-2-yl]oxy}-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl]oxy}-4-hydroxy-6-({[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}methyl)tetrahydro-2H-pyran-2-yl]oxy}-6-carbamoyl-4-(carbamoyloxy)-5-hydroxy-5-methyltetrahydro-2H-pyran-2-yl]oxy}(hydroxy)phosphoryl]oxy}-2-{[(2Z)-3,7-dimethylocta-2,6-dien-1-yl]oxy}propanoic acid, Penicillin-insensitive transglycosylase | | Authors: | Yuan, Y, Sliz, P, Walker, S. | | Deposit date: | 2008-05-11 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of the contacts anchoring moenomycin to peptidoglycan glycosyltransferases and implications for antibiotic design.
Acs Chem.Biol., 3, 2008
|
|
2ZKO
 
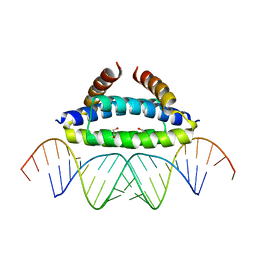 | |
3AXJ
 
 | | High resolution crystal structure of C3PO | | Descriptor: | GM27569p, Translin associated factor X, isoform B | | Authors: | Yuan, Y.A, Yang, X. | | Deposit date: | 2011-04-07 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High resolution crystal structure of C3PO
To be Published
|
|
1CF5
 
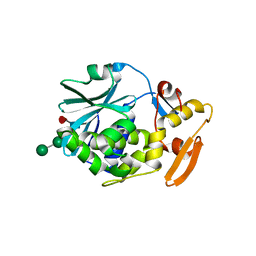 | | BETA-MOMORCHARIN STRUCTURE AT 2.55 A | | Descriptor: | PROTEIN (BETA-MOMORCHARIN), beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yuan, Y.-R, He, Y.-N, Xiong, J.-P, Xia, Z.-X. | | Deposit date: | 1999-03-24 | | Release date: | 1999-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structure of beta-momorcharin at 2.55 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5V4U
 
 | |
7WSM
 
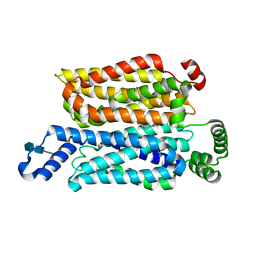 | | Cryo-EM structure of human glucose transporter GLUT4 bound to cytochalasin B in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytochalasin B, Solute carrier family 2, ... | | Authors: | Yuan, Y, Kong, F, Xu, H, Zhu, A, Yan, N, Yan, C. | | Deposit date: | 2022-01-30 | | Release date: | 2022-05-18 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Cryo-EM structure of human glucose transporter GLUT4.
Nat Commun, 13, 2022
|
|
