7U5M
 
 | | Cryo-EM Structure of GAPDH | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Morgan, C.E, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
7U5H
 
 | | Cryo-EM Structure of DNPEP | | Descriptor: | Aspartyl aminopeptidase, ZINC ION | | Authors: | Morgan, C.E, Yu, E.W, Zhang, Z. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
7U5K
 
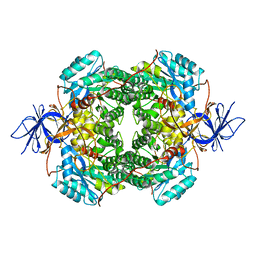 | | Cryo-EM Structure of DPYSL2 | | Descriptor: | Dihydropyrimidinase-related protein 2 | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
7U5N
 
 | |
7U5L
 
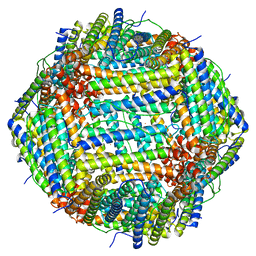 | | Cryo-EM Structure of Ferritin | | Descriptor: | FE (III) ION, Ferritin heavy chain | | Authors: | Morgan, C.E, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
7U5I
 
 | |
2HQC
 
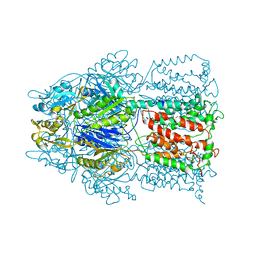 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
2HQG
 
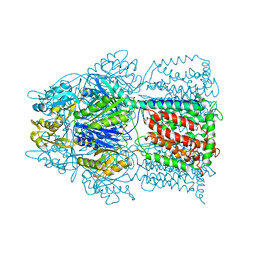 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
4R1I
 
 | |
2HQF
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
7JZ2
 
 | | Succinate: quinone oxidoreductase SQR from E.coli K12 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lyu, M, Su, C.-C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
9B43
 
 | |
9B46
 
 | |
6MNA
 
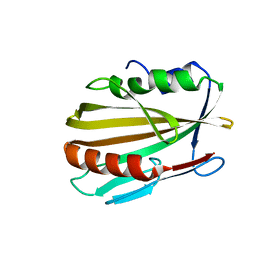 | |
3QPS
 
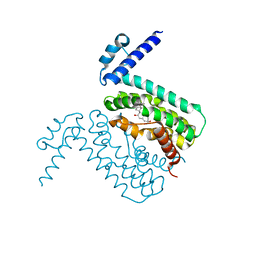 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
3QQA
 
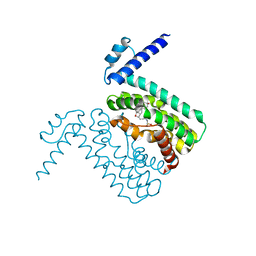 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.-C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
3T51
 
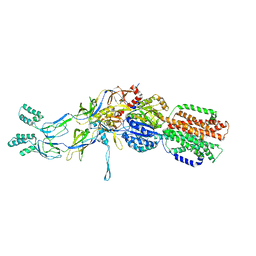 | | Crystal structures of the pre-extrusion and extrusion states of the CusBA adaptor-transporter complex | | Descriptor: | COPPER (II) ION, Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E.W. | | Deposit date: | 2011-07-26 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
3T56
 
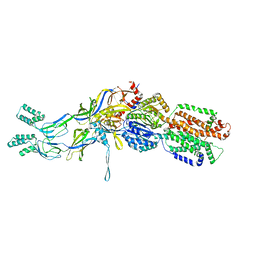 | | Crystal structure of the pre-extrusion state of the CusBA adaptor-transporter complex | | Descriptor: | COPPER (II) ION, Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E.W. | | Deposit date: | 2011-07-26 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
3T6N
 
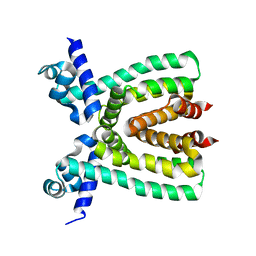 | |
3H94
 
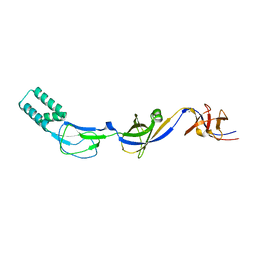 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | Descriptor: | Cation efflux system protein cusB, SILVER ION | | Authors: | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | Deposit date: | 2009-04-30 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Crystal structure of the membrane fusion protein CusB from Escherichia coli
J.Mol.Biol., 393, 2009
|
|
3T53
 
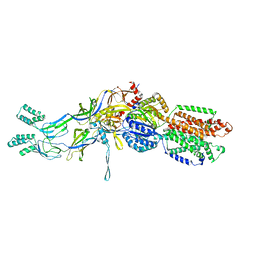 | | Crystal structures of the extrusion state of the CusBA adaptor-transporter complex | | Descriptor: | COPPER (II) ION, Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E.W. | | Deposit date: | 2011-07-26 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
7RYF
 
 | | A. baumannii Ribosome-TP-6076 complex: P-site tRNA 70S | | Descriptor: | (4S,4aS,5aR,12aS)-4-(diethylamino)-3,10,12,12a-tetrahydroxy-1,11-dioxo-8-[(2S)-pyrrolidin-2-yl]-7-(trifluoromethyl)-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide, 16S Ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-02 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | An Analysis of the Novel Fluorocycline TP-6076 Bound to Both the Ribosome and Multidrug Efflux Pump AdeJ from Acinetobacter baumannii.
Mbio, 13, 2022
|
|
4NB5
 
 | |
4MT0
 
 | |
7RYG
 
 | | A. baumannii Ribosome-TP-6076 complex: E-site tRNA 70S | | Descriptor: | (4S,4aS,5aR,12aS)-4-(diethylamino)-3,10,12,12a-tetrahydroxy-1,11-dioxo-8-[(2S)-pyrrolidin-2-yl]-7-(trifluoromethyl)-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide, 16S Ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-02 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | An Analysis of the Novel Fluorocycline TP-6076 Bound to Both the Ribosome and Multidrug Efflux Pump AdeJ from Acinetobacter baumannii.
Mbio, 13, 2022
|
|
