1IY5
 
 | | Solution structure of wild type OMSVP3 | | Descriptor: | OMSVP3 | | Authors: | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | Deposit date: | 2002-07-23 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
7CEE
 
 | | Crystal structure of mouse neuroligin-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroligin-3 | | Authors: | Yamagata, A, Yoshida, T, Shiroshima, T, Maeda, A, Fukai, S. | | Deposit date: | 2020-06-23 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.763 Å) | | Cite: | Canonical versus non-canonical transsynaptic signaling of neuroligin 3 tunes development of sociality in mice.
Nat Commun, 12, 2021
|
|
7CEG
 
 | | Crystal structure of the complex between mouse PTP delta and neuroligin-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform C of Receptor-type tyrosine-protein phosphatase delta, Neuroligin-3 | | Authors: | Yamagata, A, Yoshida, T, Shiroshima, T, Maeda, A, Fukai, S. | | Deposit date: | 2020-06-23 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Canonical versus non-canonical transsynaptic signaling of neuroligin 3 tunes development of sociality in mice.
Nat Commun, 12, 2021
|
|
2D0U
 
 | | Crystal structure of cyanide bound form of human indoleamine 2,3-dioxygenase | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CYANIDE ION, Indoleamine 2,3-dioxygenase, ... | | Authors: | Sugimoto, H, Oda, S, Otsuki, T, Hino, T, Yoshida, T, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-08 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1WKI
 
 | | solution structure of ribosomal protein L16 from thermus thermophilus HB8 | | Descriptor: | LSU ribosomal protein L16P | | Authors: | Nishimura, M, Yoshida, T, Shirouzu, M, Terada, T, Kuramitsu, S, Yokoyama, S, Ohkubo, T, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribosomal Protein L16 from Thermus thermophilus HB8
J.Mol.Biol., 344, 2004
|
|
3VJK
 
 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with MP-513 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-ylcarbonyl]thiazolidine): A highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem., 20, 2012
|
|
7CK5
 
 | | Solution structure of 28 amino acid polypeptide (354-381) in Plantago asiatica mosaic virus replicase bound to SDS micelle | | Descriptor: | PlAMV replicase peptide from RNA-dependent RNA polymerase | | Authors: | Komatsu, K, Sasaki, N, Yoshida, T, Suzuki, K, Masujima, Y, Hashimoto, M, Watanabe, S, Tochio, N, Kigawa, T, Yamaji, Y, Oshima, K, Namba, S, Nelson, R, Arie, T. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of a Proline-Kinked Amphipathic alpha-Helix Downstream from the Methyltransferase Domain of a Potexvirus Replicase and Its Role in Virus Replication and Perinuclear Complex Formation.
J.Virol., 95, 2021
|
|
1Y1C
 
 | | Solution structure of Anemonia elastase inhibitor analogue | | Descriptor: | Elastase inhibitor | | Authors: | Hemmi, H, Kumazaki, T, Yoshizawa-Kumagaye, K, Nishiuchi, Y, Yoshida, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2004-11-18 | | Release date: | 2005-07-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Study of an Anemonia Elastase Inhibitor, a "Nonclassical" Kazal-Type Inhibitor from Anemonia sulcata
Biochemistry, 44, 2005
|
|
3VJL
 
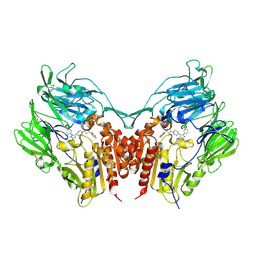 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with a prolylthiazolidine inhibitor #2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-ylcarbonyl]thiazolidine): A highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem., 20, 2012
|
|
1Y1B
 
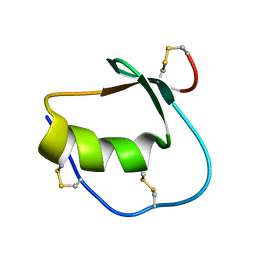 | | Solution structure of Anemonia elastase inhibitor | | Descriptor: | Elastase inhibitor | | Authors: | Hemmi, H, Kumazaki, T, Yoshizawa-Kumagaye, K, Nishiuchi, Y, Yoshida, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2004-11-18 | | Release date: | 2005-07-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Study of an Anemonia Elastase Inhibitor, a "Nonclassical" Kazal-Type Inhibitor from Anemonia sulcata
Biochemistry, 44, 2005
|
|
2D0T
 
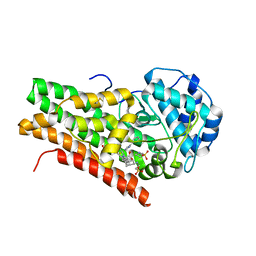 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase, ... | | Authors: | Sugimoto, H, Oda, S, Otsuki, T, Hino, T, Yoshida, T, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-08 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1WE1
 
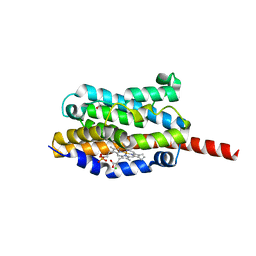 | | Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC6803 in complex with heme | | Descriptor: | CHLORIDE ION, Heme oxygenase 1, ISOPROPYL ALCOHOL, ... | | Authors: | Sugishima, M, Migita, C.T, Zhang, X, Yoshida, T, Fukuyama, K. | | Deposit date: | 2004-05-21 | | Release date: | 2004-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC 6803 in complex with heme
Eur.J.Biochem., 271, 2004
|
|
1ISE
 
 | | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly | | Descriptor: | Ribosome Recycling Factor | | Authors: | Nakano, H, Yoshida, T, Oka, S, Uchiyama, S, Nishina, K, Ohkubo, T, Kato, H, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-30 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly
To be Published
|
|
1WW9
 
 | | Crystal structure of the terminal oxygenase component of carbazole 1,9a-dioxygenase, a non-heme iron oxygenase system catalyzing the novel angular dioxygenation for carbazole and dioxin | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, terminal oxygenase component of carbazole | | Authors: | Nojiri, H, Ashikawa, Y, Noguchi, H, Nam, J.-W, Urata, M, Fujimoto, Z, Mizuno, H, Yoshida, T, Habe, H, Omori, T. | | Deposit date: | 2005-01-05 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the terminal oxygenase component of angular dioxygenase, carbazole 1,9a-dioxygenase
J.Mol.Biol., 351, 2005
|
|
1IW4
 
 | | Solution structure of ascidian trypsin inhibitor | | Descriptor: | trypsin inhibitor | | Authors: | Hemmi, H, Yoshida, T, Kumazaki, T, Nemoto, N, Hasegawa, J, Nishioka, F, Kyogoku, Y, Yokosawa, H, Kobayashi, Y. | | Deposit date: | 2002-04-19 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ascidian trypsin inhibitor determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 41, 2002
|
|
1VCK
 
 | | Crystal structure of ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10 | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, HYDROSULFURIC ACID, ... | | Authors: | Nam, J.-W, Noguchi, H, Fujiomoto, Z, Mizuno, H, Fushinobu, S, Kobashi, N, Iwata, K, Yoshida, T, Habe, H, Yamane, H, Omori, T, Nojiri, H. | | Deposit date: | 2004-03-09 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10, a novel Rieske non-heme iron oxygenase system
PROTEINS, 58, 2005
|
|
1WPK
 
 | | Methylated Form of N-terminal Transcriptional Regulator Domain of Escherichia Coli Ada Protein | | Descriptor: | ADA regulatory protein, ZINC ION | | Authors: | Takinowaki, H, Matsuda, Y, Yoshida, T, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2004-09-07 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the methylated form of the N-terminal 16-kDa domain of Escherichia coli Ada protein
Protein Sci., 15, 2006
|
|
4Y61
 
 | | Crystal structure of the complex between Slitrk2 LRR1 and PTP delta Ig1-Fn1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase delta, SLIT and NTRK-like protein 2 | | Authors: | Yamgata, A, Sato, Y, Goto-Ito, S, Uemura, T, Maeda, A, Shiroshima, T, Yoshida, T, Fukai, S. | | Deposit date: | 2015-02-12 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.358 Å) | | Cite: | Structure of Slitrk2-PTP delta complex reveals mechanisms for splicing-dependent trans-synaptic adhesion.
Sci Rep, 5, 2015
|
|
1Y69
 
 | | RRF domain I in complex with the 50S ribosomal subunit from Deinococcus radiodurans | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L16, 50S ribosomal protein L27, ... | | Authors: | Wilson, D.N, Schluenzen, F, Harms, J.M, Yoshida, T, Ohkubo, T, Albrecht, R, Buerger, J, Kobayashi, Y, Fucini, P. | | Deposit date: | 2004-12-04 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | X-ray crystallography on ribosome recycling: mechanism of binding and action of RRF on the 50S ribosomal subunit
EMBO J., 24, 2005
|
|
2E5E
 
 | | Solution Structure of Variable-type Domain of Human Receptor for Advanced Glycation Endproducts | | Descriptor: | Advanced glycosylation end product-specific receptor | | Authors: | Matsumoto, S, Yoshida, T, Yasumatsu, I, Yamamoto, H, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Variable-type Domain of Human Receptor for Advanced Glycation Endproducts
to be published
|
|
1VA2
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 2) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1V8X
 
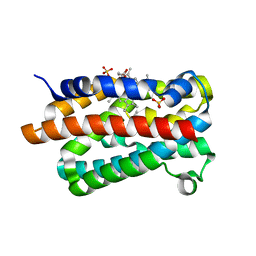 | | Crystal Structure of the Dioxygen-bound Heme Oxygenase from Corynebacterium diphtheriae | | Descriptor: | Heme oxygenase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Unno, M, Matsui, T, Chu, G.C, Couture, M, Yoshida, T, Rousseau, D.L, Olson, J.S, Ikeda-Saito, M. | | Deposit date: | 2004-01-15 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Dioxygen-bound Heme Oxygenase from Corynebacterium diphtheriae: IMPLICATIONS FOR HEME OXYGENASE FUNCTION.
J.Biol.Chem., 279, 2004
|
|
1V6R
 
 | | Solution Structure of Endothelin-1 with its C-terminal Folding | | Descriptor: | Endothelin-1 | | Authors: | Takashima, H, Mimura, N, Ohkubo, T, Yoshida, T, Tamaoki, H, Kobayashi, Y. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Distributed Computing and NMR Constraint-Based High-Resolution Structure
Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal
Folding
J.Am.Chem.Soc., 126, 2004
|
|
1VA3
 
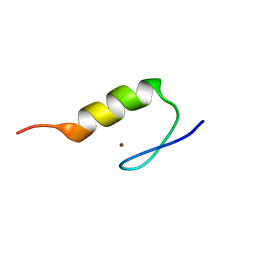 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 3) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1VA1
 
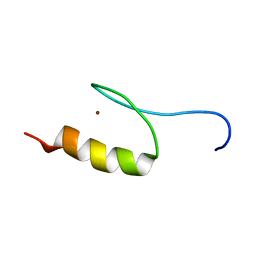 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 1) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
