1WW4
 
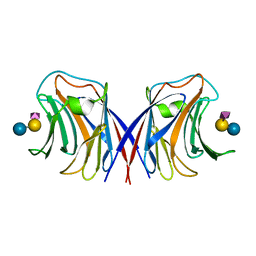 | | Agrocybe cylindracea galectin complexed with NeuAca2-3lactose | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-alpha-D-galactopyranose-(1-4)-alpha-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin | | Authors: | Ban, M, Yoon, H.J, Demirkan, E, Utsumi, S, Mikami, B, Yagi, F. | | Deposit date: | 2004-12-31 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of a Fungal Galectin from Agrocybe cylindracea for Recognizing Sialoconjugate
J.Mol.Biol., 351, 2005
|
|
1WW5
 
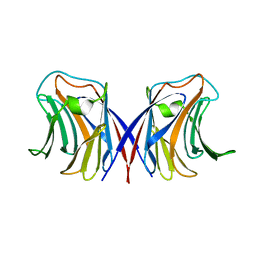 | | Agrocybe cylindracea galectin complexed with 3'-sulfonyl lactose | | Descriptor: | 3-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin | | Authors: | Ban, M, Yoon, H.J, Demirkan, E, Utsumi, S, Mikami, B, Yagi, F. | | Deposit date: | 2005-01-03 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of a Fungal Galectin from Agrocybe cylindracea for Recognizing Sialoconjugate
J.Mol.Biol., 351, 2005
|
|
1WW7
 
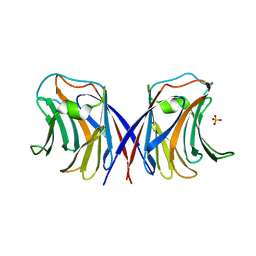 | | Agrocybe cylindracea galectin (Ligand-free) | | Descriptor: | SULFATE ION, galectin | | Authors: | Ban, M, Yoon, H.J, Demirkan, E, Utsumi, S, Mikami, B, Yagi, F. | | Deposit date: | 2005-01-03 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of a Fungal Galectin from Agrocybe cylindracea for Recognizing Sialoconjugate
J.Mol.Biol., 351, 2005
|
|
4OID
 
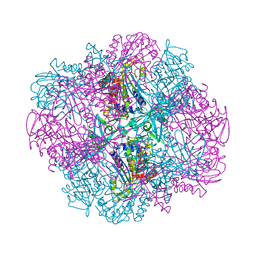 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | Probable M18 family aminopeptidase 2 | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.D, Ha, S.C, Yoon, H.J, Kim, K.S, Yun, K.H, Kim, J.H, Kim, K.K. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4Q9D
 
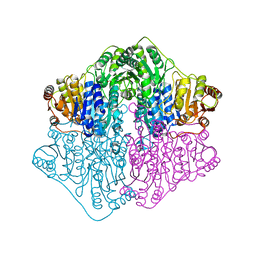 | | X-ray structure of a putative thiamin diphosphate-dependent enzyme isolated from Mycobacterium smegmatis | | Descriptor: | Benzoylformate decarboxylase, FORMIC ACID, MAGNESIUM ION | | Authors: | Andrews, F.H, Horton, J.D, Yoon, H.J, Malik, A.M.K, Logsdon, M.G, Shin, D.H, Kneen, M.M, Suh, S.W, McLeish, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The kinetic characterization and X-ray structure of a putative benzoylformate decarboxylase from M. smegmatis highlights the difficulties in the functional annotation of ThDP-dependent enzymes.
Biochim.Biophys.Acta, 1854, 2015
|
|
4Q6Q
 
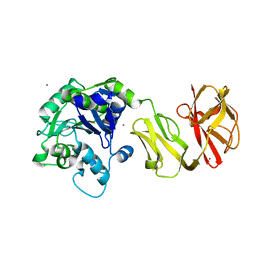 | | Structural analysis of the Zn-form II of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6N
 
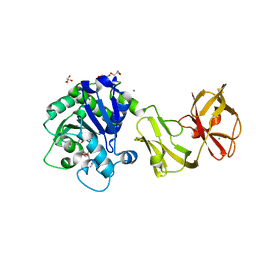 | | Structural analysis of the tripeptide-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6M
 
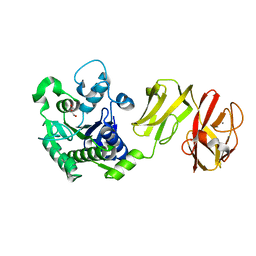 | | Structural analysis of the apo-form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6O
 
 | | Structural analysis of the mDAP-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OIW
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | Probable M18 family aminopeptidase 2, ZINC ION | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.D, Ha, S.C, Yoon, H.J, Kim, K.S, Yun, K.H, Kim, J.H, Kim, K.K. | | Deposit date: | 2014-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4Q6P
 
 | | Structural analysis of the Zn-form I of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QB9
 
 | | Crystal structure of Mycobacterium smegmatis Eis in complex with paromomycin | | Descriptor: | Enhanced intracellular survival protein, PAROMOMYCIN, SULFATE ION | | Authors: | Kim, K.H, Ahn, D.R, Yoon, H.J, Yang, J.K, Suh, S.W. | | Deposit date: | 2014-05-06 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Structure of Mycobacterium smegmatis Eis in complex with paromomycin.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
5YU4
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5Z2W
 
 | | Crystal structure of the bacterial cell division protein FtsQ and FtsB | | Descriptor: | Cell division protein FtsB, Cell division protein FtsQ, MAGNESIUM ION | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the FtsQ/FtsB/FtsL Complex, a Key Component of the Divisome.
Sci Rep, 8, 2018
|
|
5YU0
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5XE3
 
 | | Endoribonuclease in complex with its cognate antitoxin from Mycobacterial species | | Descriptor: | Endoribonuclease MazF4, Probable antitoxin MazE4 | | Authors: | Ahn, D.-H, Lee, K.-Y, Lee, S.J, Yoon, H.J, Kim, S.-J, Lee, B.-J. | | Deposit date: | 2017-03-31 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of the MazEF4 toxin-antitoxin pair in Mycobacterium tuberculosis provide evidence for a unique extracellular death factor.
J. Biol. Chem., 292, 2017
|
|
5YU1
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | (2S)-piperidine-2-carboxylic acid, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5X3T
 
 | | VapBC from Mycobacterium tuberculosis | | Descriptor: | Antitoxin VapB26, MAGNESIUM ION, Ribonuclease VapC26 | | Authors: | Kang, S.M, Kim, D.H, Yoon, H.J, Lee, B.J. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-07 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional details of the Mycobacterium tuberculosis VapBC26 toxin-antitoxin system based on a structural study: insights into unique binding and antibiotic peptides.
Nucleic Acids Res., 45, 2017
|
|
5YU3
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROLINE, ... | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5XE2
 
 | | Endoribonuclease from Mycobacterial species | | Descriptor: | Endoribonuclease MazF4 | | Authors: | Ahn, D.-H, Lee, K.-Y, Lee, S.J, Yoon, H.J, Kim, S.-J, Lee, B.-J. | | Deposit date: | 2017-03-31 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural analyses of the MazEF4 toxin-antitoxin pair in Mycobacterium tuberculosis provide evidence for a unique extracellular death factor.
J. Biol. Chem., 292, 2017
|
|
